Quasi-species prevalence and clinical impact of evolving SARS-CoV-2 lineages in European COVID-19 cohorts, January 2020 to February 2022
- PMID: 40084424
- PMCID: PMC11912139
- DOI: 10.2807/1560-7917.ES.2025.30.10.2400038
Quasi-species prevalence and clinical impact of evolving SARS-CoV-2 lineages in European COVID-19 cohorts, January 2020 to February 2022
Abstract
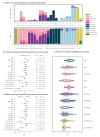
BackgroundEvolution of SARS-CoV-2 is continuous.AimBetween 01/2020 and 02/2022, we studied SARS-CoV-2 variant epidemiology, evolution and association with COVID-19 severity.MethodsIn nasopharyngeal swabs of COVID-19 patients (n = 1,762) from France, Italy, Spain, and the Netherlands, SARS-CoV-2 was investigated by reverse transcription-quantitative PCR and whole-genome sequencing, and the virus variant/lineage (NextStrain/Pangolin) was determined. Patients' demographic and clinical details were recorded. Associations between mild/moderate or severe COVID-19 and SARS-CoV-2 variants and patient characteristics were assessed by logistic regression. Rates and genomic locations of mutations, as well as quasi-species distribution (≥ 2 heterogeneous positions, ≥ 50× coverage) were estimated based on 1,332 high-quality sequences.ResultsOverall, 11 SARS-CoV-2 clades infected 1,762 study patients of median age 59 years (interquartile range (IQR): 45-73), with 52.5% (n = 925) being male. In total, 101 non-synonymous substitutions/insertions correlated with disease prognosis (severe, n = 27; mild-to-moderate, n = 74). Several hotspots (mutation rates ≥ 85%) occurred in Alpha, Delta, and Omicron variants of concern (VOCs) but none in pre-Alpha strains. Four hotspots were retained across all study variants, including spike:D614G. Average number of mutations per open-reading-frame (ORF) increased in the spike gene (average < 5 per genome in January 2020 to > 15 in 2022), but remained stable in ORF1ab, membrane, and nucleocapsid genes. Quasi-species were most prevalent in 20A/EU2 (48.9%), 20E/EU1 (48.6%), 20A (38.8%), and 21K/Omicron (36.1%) infections. Immunocompromised status and age (≥ 60 years), while associated with severe COVID-19 or death irrespective of variant (odds ratio (OR): 1.60-2.25; p ≤ 0.014), did not affect quasi-species' prevalence (p > 0.05).ConclusionSpecific mutations correlate with COVID-19 severity. Quasi-species potentially shaping VOCs' emergence are relevant to consider.
Keywords: France; Genome evolution; Italy; ORCHESTRA; RNA secondary structure; SNV; Spain; The Netherlands; coronavirus; deletions; insertions; mutation rate; mutations; quasi-species.
Conflict of interest statement
Figures




References
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
