CD103+CD56+ ILCs Are Associated with an Altered CD8+ T-cell Profile within the Tumor Microenvironment
- PMID: 40084939
- PMCID: PMC11962407
- DOI: 10.1158/2326-6066.CIR-24-0151
CD103+CD56+ ILCs Are Associated with an Altered CD8+ T-cell Profile within the Tumor Microenvironment
Abstract
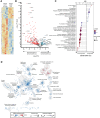
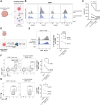
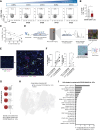
Immunotherapies have had unprecedented success in the treatment of multiple cancer types, albeit with variable response rates. Unraveling the complex network of immune cells within the tumor microenvironment (TME) may provide additional insights to enhance antitumor immunity and improve clinical response. Many studies have shown that NK cells or innate lymphoid cells (ILC) have regulatory capacity. Here, we identified CD103 as a marker that was found on CD56+ cells that were associated with a poor proliferative capacity of tumor-infiltrating lymphocytes in culture. We further demonstrated that CD103+CD56+ ILCs isolated directly from tumors represented a distinct ILC population that expressed unique surface markers (such as CD49a and CD101), transcription factor networks, and transcriptomic profiles compared with CD103-CD56+ NK cells. Using single-cell multiomic and spatial approaches, we found that these CD103+CD56+ ILCs were associated with CD8+ T cells with reduced expression of granzyme B. Thus, this study identifies a population of CD103+CD56+ ILCs with potentially inhibitory functions that are associated with a TME that includes CD8+ T cells with poor antitumor activity. Further studies focusing on these cells may provide additional insights into the biology of an inhibitory TME.
©2025 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
H.W. Jackson reports grants, personal fees, and nonfinancial support from Standard BioTools, personal fees and nonfinancial support from Abcam, and nonfinancial support from Somalogic outside the submitted work. P.S Ohashi reports grants from Canadian Institutes of Health Research, Canadian Cancer Society, TRANSCAN3, and Princess Margaret Cancer Foundation Wolfond Immunotherapy Fund during the conduct of the study, as well as other support from Providence Therapeutics, Treadwell Therapeutics, Tikvo Allocell, and Rondo Therapeutics outside the submitted work. S.Q. Crome, P.S. Ohashi, and L.T. Nguyen filed a patent serial no. 16/325,923. No disclosures were reported by the other authors.
Figures




References
-
- Vivier E, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, et al. . Innate lymphoid cells: 10 years on. Cell 2018;174:1054–66. - PubMed
-
- Bernink JH, Krabbendam L, Germar K, de Jong E, Gronke K, Kofoed-Nielsen M, et al. . Interleukin-12 and -23 control plasticity of CD127+ group 1 and group 3 innate lymphoid cells in the intestinal lamina propria. Immunity 2015;43:146–60. - PubMed
-
- Crinier A, Kerdiles Y, Vienne M, Cózar B, Vivier E, Berruyer C. Multidimensional molecular controls defining NK/ILC1 identity in cancers. Semin Immunol 2021;52:101424. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

