Deep Learning Radiopathomics Models Based on Contrast-enhanced MRI and Pathologic Imaging for Predicting Vessels Encapsulating Tumor Clusters and Prognosis in Hepatocellular Carcinoma
- PMID: 40084948
- PMCID: PMC11966553
- DOI: 10.1148/rycan.240213
Deep Learning Radiopathomics Models Based on Contrast-enhanced MRI and Pathologic Imaging for Predicting Vessels Encapsulating Tumor Clusters and Prognosis in Hepatocellular Carcinoma
Abstract
Purpose To develop deep learning (DL) radiopathomics models based on contrast-enhanced MRI and pathologic imaging to predict vessels encapsulating tumor clusters (VETC) and survival in hepatocellular carcinoma (HCC). Materials and Methods In this retrospective, multicenter study, 578 patients with HCC (mean age [±SD], 59 years ± 10; 442 male, 136 female) were divided into the training (n = 317), internal (n = 137), and external (n = 124) test sets. DL radiomics and pathomics models were developed to predict VETC using gadoxetic acid-enhanced MR and pathologic images. Deep radiomics score (DRS) and handcrafted and deep pathomics scores were compared between the group with VETC pattern in HCC (VETC+) and group without VETC pattern in HCC (VETC-). Multivariable Cox regression analyses were performed to identify independent prognostic factors, and the radiopathomics nomogram models were developed for early recurrence and progression-free survival (PFS). The prognostic power was evaluated using the concordance index (C index) and time-dependent receiver operating characteristic (ROC) curves. Results In the external test set, the Swin Transformer showed good performance for predicting VETC in both DL radiomics (area under the ROC curve [AUC], 0.77-0.79) and pathomics (AUC, 0.79) models. Patients with VETC+ HCC had significantly higher DRS and handcrafted and deep pathomics scores compared with patients with VETC- HCC in all datasets (all P < .001). The radiopathomics nomogram model incorporating DRS in the arterial phase and the handcrafted and deep pathomics scores achieved C indexes of 0.69, 0.60, and 0.67 for early recurrence and time-dependent AUCs of 0.83 (95% CI: 0.76, 0.91), 0.81 (95% CI: 0.68, 0.94), and 0.78 (95% CI: 0.67, 0.88) for 3-year PFS in the training, internal, and external test sets, respectively. Early recurrence and PFS rates statistically significantly differed between the high- and low-risk patients stratified by the radiopathomics nomogram model (all P < .05). Conclusion DL radiopathomics models effectively helped to predict VETC in HCC and assess the risk for early recurrence and PFS. Keywords: Hepatocellular Carcinoma, Deep Learning, MRI, Radiopathomics, Survival Supplemental material is available for this article. © RSNA, 2025.
Keywords: Deep Learning; Hepatocellular Carcinoma; MRI; Radiopathomics; Survival.
Conflict of interest statement
Figures

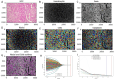


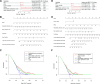

Comment in
-
Deep Learning Radiopathomics for Predicting Tumor Vasculature and Prognosis in Hepatocellular Carcinoma.Radiol Imaging Cancer. 2025 May;7(3):e250141. doi: 10.1148/rycan.250141. Radiol Imaging Cancer. 2025. PMID: 40314587 Free PMC article. No abstract available.
References
-
- Feng Z , Li H , Zhao H , et al. . Preoperative CT for characterization of aggressive macrotrabecular-massive subtype and vessels that encapsulate tumor clusters pattern in hepatocellular carcinoma . Radiology 2021. ; 300 ( 1 ): 219 – 229 . - PubMed
-
- Fang JH , Zhou HC , Zhang C , et al. . A novel vascular pattern promotes metastasis of hepatocellular carcinoma in an epithelial-mesenchymal transition-independent manner . Hepatology 2015. ; 62 ( 2 ): 452 – 465 . - PubMed
-
- Renne SL , Woo HY , Allegra S , et al. . Vessels encapsulating tumor clusters (VETC) is a powerful predictor of aggressive hepatocellular carcinoma . Hepatology 2020. ; 71 ( 1 ): 183 – 195 . - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

