Aged and BRCA-Mutated Stromal Cells Drive Epithelial Cell Transformation
- PMID: 40084985
- PMCID: PMC12130807
- DOI: 10.1158/2159-8290.CD-24-0805
Aged and BRCA-Mutated Stromal Cells Drive Epithelial Cell Transformation
Abstract
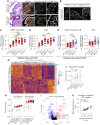
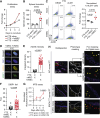
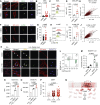
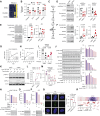
The fundamental steps in high-grade serous ovarian cancer (HGSOC) initiation are unclear, presenting critical barriers to the prevention and early detection of this deadly disease. Current models propose that fallopian tube epithelial (FTE) cells transform into serous tubal intraepithelial carcinoma (STIC) precursor lesions and subsequently into HGSOC. In this study, we report that an epigenetically altered mesenchymal stem cell niche, termed high-risk mesenchymal stromal/stem cell (hrMSC), exists prior to STIC lesion formation. hrMSCs are enriched in STIC stroma and contribute to a stromal "field effect" extending beyond the borders of the STIC lesion. hrMSCs promote DNA damage in FTE cells while also fostering FTE cell survival. hrMSCs induce malignant transformation of the FTE, resulting in metastatic cancer in vivo, indicating that hrMSCs promote cancer initiation. hrMSCs are significantly enriched in BRCA1/2 mutation carriers and increase with age. Combined, these findings indicate that hrMSCs can incite ovarian cancer initiation and have important implications for ovarian cancer detection and prevention.
Significance: This work demonstrates a critical role of fallopian tube stromal cells in HGSOC initiation with implications for the pathophysiology of HGSOC formation and the development of prevention and early detection strategies critically needed in this disease. Additionally, the identification of stromal-mediated epithelial transformation has broad implications for understanding pan-cancer initiation. See related commentary by Recouvreux and Orsulic, p. 1093.
©2025 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
I.P. MacFawn reports grants from the NIH during the conduct of the study as well as personal fees from Galvanize Therapeutics, Inc. outside the submitted work. N. Hempel reports grants from the NIH and Department of Defense (DoD) during the conduct of the study. R. Bao reports other support from the University of Chicago (ClostraBio) outside the submitted work. R. Drapkin reports personal fees from Repare Therapeutics, Light Horse Therapeutics, and ImmunoGen, Inc. outside the submitted work. L.G. Coffman reports grants from the NIH, Tina’s Wish, and the Department of Defense during the conduct of the study. No disclosures were reported by the other authors.
Figures



References
-
- Siegel RL, Giaquinto AN, Jemal A. Cancer statistics, 2024. CA Cancer J Clin 2024;74:12–49. - PubMed
MeSH terms
Substances
Grants and funding
- Honorable Tina Brozman Foundation (Tina's Wish)
- R01CA242021/National Cancer Institute (NCI)
- P50 CA272218/CA/NCI NIH HHS/United States
- P30 CA047904/CA/NCI NIH HHS/United States
- S10OD028483/National Cancer Institute (NCI)
- OC210139/U.S. Department of Defense (DOD)
- Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (AMRF)
- AG077923/National Cancer Institute (NCI)
- R01 CA242021/CA/NCI NIH HHS/United States
- P50 CA228991/CA/NCI NIH HHS/United States
- S10 OD028483/OD/NIH HHS/United States
- W81XWH-22-1-0852/U.S. Department of Defense (DOD)
- U01 AG077923/AG/NIA NIH HHS/United States
- P50CA272218-01A1/National Cancer Institute (NCI)
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

