HLA-E/peptide complexes differentially interact with NKG2A/CD94 and T cell receptors
- PMID: 40085431
- PMCID: PMC12041767
- DOI: 10.1093/jimmun/vkae068
HLA-E/peptide complexes differentially interact with NKG2A/CD94 and T cell receptors
Abstract
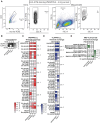
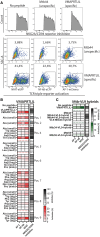
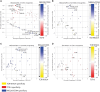
The virtually monomorphic antigen presentation molecule HLA-E can present self- and non-self peptides to the NKG2A/CD94 co-receptor inhibitory complex expressed on natural killer (NK) cells and to T cell receptors (TCRs) expressed on T cells. HLA-E presents self-peptides to NKG2A/CD94 to regulate tissue homeostasis, whereas HLA-E restricted T cells mediate regulatory and cytotoxic responses toward pathogen-infected cells. In this study, we directly compared HLA-E/peptide recognition and signaling between NKG2A/CD94 and 2 HLA-E restricted TCRs that can recognize self-peptides or identical peptide mimics from the viral UL40 protein of cytomegalovirus using position substituted peptide variants. We show that position 7 is critical for interaction with NKG2A/CD94, whereas position 8 is important for interaction with the TCRs. The Arginine at position 5 of these peptides is an essential residue for recognition by both receptors. Thus, NKG2A/CD94 and TCRs have different requirements for recognition of peptides presented in HLA-E.
Keywords: HLA-E; NKG2A/CD94; T cell receptor; peptide/receptor interactions.
© The Author(s) 2025. Published by Oxford University Press on behalf of The American Association of Immunologists.
Conflict of interest statement
None declared.
Figures

References
-
- Strong RK et al. HLA-E allelic variants. Correlating differential expression, peptide affinities, crystal structures, and thermal stabilities. J Biol Chem. 2003;278:5082–5090. - PubMed
-
- Grimsley C, Ober C. Population genetic studies of HLA-E: evidence for selection. Hum Immunol. 1997;52:33–40. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

