Ectopic expression of testis-specific transcription elongation factor in driving cancer
- PMID: 40085698
- PMCID: PMC11908497
- DOI: 10.1126/sciadv.ads4200
Ectopic expression of testis-specific transcription elongation factor in driving cancer
Abstract
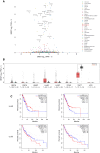
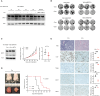
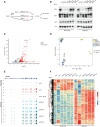
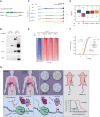
The testis-specific BET protein BRDT structurally resembles the ubiquitous BRD4 and is misexpressed in cancer, and we show that BRDT misexpression may affect lung cancer progression. BRDT knockdown in lung cancer cells slowed tumor growth and prolonged survival in a xenograft model. Comparative characterization of PTEFb complex participation and chromatin binding indicates BRD4-redundant and BRD4-distinct BRDT functions. Unlike dual depletion, individual BRD4 or BRDT knockdown did not impair transcriptional responses to hypoxia in BRDT-expressing cells, consistent with redundant function. However, BRD4 depletion/BRDT complementation revealed that BRDT can also release paused RNA polymerase II independently of its bromodomains as we previously demonstrated not to be required for Pol II pause/release function of BRD4, underscoring the functional importance of the C-terminal domains in both BRD4 and BRDT and their potential as therapeutic targets in solid tumors. Based on this study, future investigations should explore BRD4-distinct BRDT functions and BRDT misexpression driving cancer pathogenesis.
Figures

References
-
- Chen F. X., Smith E. R., Shilatifard A., Born to run: Control of transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 19, 464–478 (2018). - PubMed
-
- Aoi Y., Shilatifard A., Transcriptional elongation control in developmental gene expression, aging, and disease. Mol. Cell 83, 3972–3999 (2023). - PubMed
-
- Lin C., Smith E. R., Takahashi H., Lai K. C., Martin-Brown S., Florens L., Washburn M. P., Conaway J. W., Conaway R. C., Shilatifard A., AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol. Cell 37, 429–437 (2010). - PMC - PubMed
-
- Luo Z., Lin C., Shilatifard A., The super elongation complex (SEC) family in transcriptional control. Nat. Rev. Mol. Cell Biol. 13, 543–547 (2012). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

