Structure of a putative terminal amidation domain in natural product biosynthesis
- PMID: 40086440
- PMCID: PMC12048289
- DOI: 10.1016/j.str.2025.02.005
Structure of a putative terminal amidation domain in natural product biosynthesis
Abstract
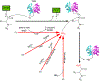
Bacteria are rich sources of pharmaceutically valuable natural products, many crafted by modular polyketide synthases (PKS) and non-ribosomal peptide synthetases (NRPS). PKS and NRPS systems typically contain a thioesterase (TE) to offload a linear or cyclized product from a carrier protein, but alternative chemistry is needed for products with a terminal amide. Several pathways with amidated products also possess an uncharacterized 400-amino acid terminal domain. We present the characterization and structure of this putative terminal amidation domain (TAD). TAD binds NAD with the nicotinamide near an invariant cysteine that is also accessible to an intermediate on a carrier protein, indicating a catalytic role. The TAD structure resembles cyanobacterial acyl-ACP reductase (AAR), which binds NADPH near an analogous catalytic cysteine. Bioinformatic analysis reveals that TADs are broadly distributed across bacterial phyla and often occur at the end of terminal NRPS modules, suggesting many amidated products may yet be discovered.
Keywords: biosynthesis; crosslinking; crystallography; genome mining; mass spectrometry; natural products.
Copyright © 2025 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures







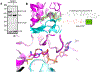
Update of
-
Structure of a Putative Terminal Amidation Domain in Natural Product Biosynthesis.bioRxiv [Preprint]. 2024 Oct 28:2024.10.28.620694. doi: 10.1101/2024.10.28.620694. bioRxiv. 2024. Update in: Structure. 2025 May 1;33(5):935-947.e4. doi: 10.1016/j.str.2025.02.005. PMID: 39554124 Free PMC article. Updated. Preprint.
References
-
- Wohlleben W, Stegmann E, and Süssmuth RD (2009). Molecular Genetic Approaches to Analyze Glycopeptide Biosynthesis. In Methods in Enzymology Complex Enzymes in Microbial Natural Product Biosynthesis, Part A: Overview Articles and Peptides. (Academic Press; ), pp. 459–486. 10.1016/S0076-6879(09)04818-6. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

