Single-cell multiomic comparison of mouse and rat spermatogenesis reveals gene regulatory networks conserved for over 20 million years
- PMID: 40086448
- PMCID: PMC12069898
- DOI: 10.1016/j.stemcr.2025.102449
Single-cell multiomic comparison of mouse and rat spermatogenesis reveals gene regulatory networks conserved for over 20 million years
Abstract
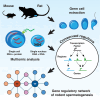
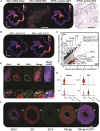
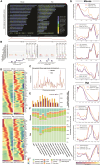
Spermatogenesis is driven by dramatic changes in chromatin regulation, gene transcription, and protein expression. To assess the mechanistic bases for these developmental changes, we utilized multiomic single-cell/nucleus RNA sequencing (sc/snRNA-seq) and single-nucleus assay for transposase-accessible chromatin with sequencing (snATAC-seq) to identify chromatin changes associated with transcription in adult mouse and rat testes. We characterized the relationships between the transcriptomes and chromatin of both species, including the divergent expression of Id4 in spermatogonial stem cells between species. Promoter accessibility and gene expression showed the greatest association during meiosis in both species. We mapped the cross-species conservation of putative regulatory regions for key spermatogenic genes, including Cd9 and Spam1, and investigated correlations and disconnects in chromatin accessibility, gene expression, and protein expression via antibody-derived tags. Using a gene regulatory network (GRN) model, we identified 40 core regulons conserved between mouse and rat germ cells, highlighting the relevance of chromatin-related factors in regulating the transcription of canonical genes across spermatogenesis.
Keywords: ATAC; RNA sequencing; differentiation; germ cells; mouse; multiomic; rat; single-cell; spermatogenesis.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





References
-
- Barroca V., Racine C., Pays L., Fouchet P., Coureuil M., Allemand I. The netrin-1 receptor UNC5C contributes to the homeostasis of undifferentiated spermatogonia in adult mice. Stem Cell Res. 2022;60 - PubMed
-
- Boussouar F., Benahmed M. Lactate and energy metabolism in male germ cells. Trends Endocrinol. Metabol. 2004;15:345–350. - PubMed
-
- Buaas F.W., Kirsh A.L., Sharma M., McLean D.J., Morris J.L., Griswold M.D., de Rooij D.G., Braun R.E. Plzf is required in adult male germ cells for stem cell self-renewal. Nat. Genet. 2004;36:647–652. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

