MdfA is a novel ClpC adaptor protein that functions in the developing Bacillus subtilis spore
- PMID: 40086879
- PMCID: PMC11960690
- DOI: 10.1101/gad.352498.124
MdfA is a novel ClpC adaptor protein that functions in the developing Bacillus subtilis spore
Abstract
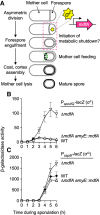
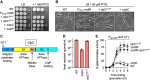
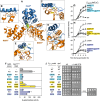
Bacterial protein degradation machinery consists of chaperone-protease complexes that play vital roles in bacterial growth and development and have sparked interest as novel antimicrobial targets. ClpC-ClpP (ClpCP) is one such chaperone-protease complex, recruited by adaptors to specific functions in the model bacterium Bacillus subtilis and other Gram-positive bacteria, including the pathogens Staphylococcus aureus and Mycobacterium tuberculosis Here we have identified a new ClpCP adaptor protein, MdfA (metabolic differentiation factor A; formerly YjbA), in a genetic screen for factors that help drive B. subtilis toward metabolic dormancy during spore formation. A knockout of mdfA stimulates gene expression in the developing spore, while aberrant expression of mdfA during vegetative growth is toxic. MdfA binds directly to ClpC to induce its oligomerization and ATPase activity, and this interaction is required for the in vivo effects of mdfA Finally, a cocrystal structure reveals that MdfA binds to the ClpC N-terminal domain at a location analogous to that on the M. tuberculosis ClpC1 protein where bactericidal cyclic peptides bind. Altogether, our data and that of an accompanying study by Riley and colleagues support a model in which MdfA induces ClpCP-mediated degradation of metabolic enzymes in the developing spore, helping drive it toward metabolic dormancy.
Keywords: AAA+ proteases; Bacillus subtilis; ClpC; ClpCP; MdfA; X-ray crystallography; YjbA; adaptor; protein degradation; sporulation.
© 2025 Massoni et al.; Published by Cold Spring Harbor Laboratory Press.
Figures




References
Grants and funding
LinkOut - more resources
Full Text Sources
