Expression of ENL YEATS domain tumor mutations in nephrogenic or stromal lineage impairs kidney development
- PMID: 40087269
- PMCID: PMC11909213
- DOI: 10.1038/s41467-025-57926-z
Expression of ENL YEATS domain tumor mutations in nephrogenic or stromal lineage impairs kidney development
Abstract
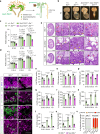
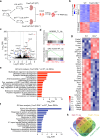
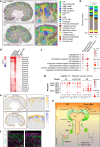
Recurrent gain-of-function mutations in the histone reader protein ENL have been identified in Wilms tumor, the most prevalent pediatric kidney cancer. However, their pathological significance in kidney development and tumorigenesis in vivo remains elusive. Here, we generate mouse models mimicking ENL tumor (ENLT) mutations and show that heterozygous mutant expression in Six2+ nephrogenic or Foxd1+ stromal lineages leads to severe, lineage-specific kidney defects, both resulting in neonatal lethality. Six2-ENLT mutant kidneys display compromised cap mesenchyme, scant nephron tubules, and cystic glomeruli, indicative of premature progenitor commitment and blocked differentiation. Bulk and spatial transcriptomic analyses reveal aberrant activation of Hox and Wnt signaling genes in mutant nephrogenic cells. In contrast, Foxd1-ENLT mutant kidneys exhibit expansion in renal capsule and cap mesenchyme, with dysregulated stromal gene expression affecting stroma-epithelium crosstalk. Our findings uncover distinct pathways through which ENL mutations disrupt nephrogenesis, providing a foundation for further investigations into their role in tumorigenesis.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

