Deep representation learning for clustering longitudinal survival data from electronic health records
- PMID: 40087274
- PMCID: PMC11909183
- DOI: 10.1038/s41467-025-56625-z
Deep representation learning for clustering longitudinal survival data from electronic health records
Abstract
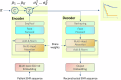
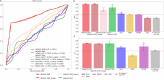
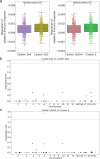
Precision medicine requires accurate identification of clinically relevant patient subgroups. Electronic health records provide major opportunities for leveraging machine learning approaches to uncover novel patient subgroups. However, many existing approaches fail to adequately capture complex interactions between diagnosis trajectories and disease-relevant risk events, leading to subgroups that can still display great heterogeneity in event risk and underlying molecular mechanisms. To address this challenge, we implemented VaDeSC-EHR, a transformer-based variational autoencoder for clustering longitudinal survival data as extracted from electronic health records. We show that VaDeSC-EHR outperforms baseline methods on both synthetic and real-world benchmark datasets with known ground-truth cluster labels. In an application to Crohn's disease, VaDeSC-EHR successfully identifies four distinct subgroups with divergent diagnosis trajectories and risk profiles, revealing clinically and genetically relevant factors in Crohn's disease. Our results show that VaDeSC-EHR can be a powerful tool for discovering novel patient subgroups in the development of precision medicine approaches.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
-
- Electronic Public Health Reporting. ONC Annu. Meet. https://www.healthit.gov/sites/default/files/2018-12/ElectronicPublicHea... (2018).
-
- Parasrampuria, S. & Henry, J. Hospitals use of electronic health records data, 2015–2017. In ASTP Health IT Data Brief [Internet] 46 (Office of the Assistant Secretary for Technology Policy, Washington, DC, 2019). - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

