Generation of hypoimmunogenic universal iPS cells through HLA-type gene knockout
- PMID: 40087529
- PMCID: PMC11958689
- DOI: 10.1038/s12276-025-01422-3
Generation of hypoimmunogenic universal iPS cells through HLA-type gene knockout
Abstract
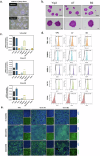
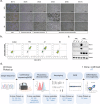
Hypoimmunogenic universal induced pluripotent stemn (iPS) cells were generated through the targeted disruption of key genes, including human leukocyte antigen (HLA)-A, HLA-B and HLA-DR alpha (DRA), using the CRISPR-Cas9 system. This approach aimed to minimize immune recognition and enhance the potential of iPS cells for allogeneic therapy. Heterozygous iPS cells were used for guide RNA design and validation to facilitate the knockout (KO) of the HLA-A, HLA-B and HLA-DRA genes. The electroporation of iPS cells using the selected guide RNAs enabled the generation of triple-KO iPS cells, followed by single-cell cloning for clone selection. Clone A7, an iPS cell with targeted KOs of the HLA-A, HLA-B and HLA-DRA genes, was identified as the final candidate. Messenger RNA analysis revealed robust expression of pluripotency markers, such as octamer-binding transcription factor 4, sex-determining region Y box 2, Krüppel-like factor 4, Lin-28 homolog A and Nanog homeobox, while protein expression assays confirmed the presence of octamer-binding transcription factor 4, stage-specific embryonic antigen 4, Nanog homeobox and tumor rejection antigen 1-60. A karyotype examination revealed no anomalies, and three-germ layer differentiation assays confirmed the differentiation potential. After interferon gamma stimulation, the gene-corrected clone A7 lacked HLA-A, HLA-B and HLA-DR protein expression. Immunogenicity testing further confirmed the hypoimmunogenicity of clone A7, which was evidenced by the absence of proliferation in central memory T cells and effector memory T cells. In conclusion, clone A7, a triple-KO iPS cell clone that demonstrates immune evasion properties, retained its intrinsic iPS cell characteristics and exhibited no immunogenicity.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





References
-
- Takahashi, K. et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell131, 861–872 (2007). - PubMed
-
- Bellin, M., Marchetto, M. C., Gage, F. H. & Mummery, C. L. Induced pluripotent stem cells: the new patient? Nat. Rev. Mol. Cell Biol.13, 713–726 (2012). - PubMed
-
- Montgomery, R. A., Tatapudi, V. S., Leffell, M. S. & Zachary, A. A. HLA in transplantation. Nat. Rev. Nephrol.14, 558–570 (2018). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

