Nasal pathobiont abundance is a moderate feedlot-dependent indicator of bovine respiratory disease in beef cattle
- PMID: 40087791
- PMCID: PMC11909826
- DOI: 10.1186/s42523-025-00387-y
Nasal pathobiont abundance is a moderate feedlot-dependent indicator of bovine respiratory disease in beef cattle
Abstract
Background: Bovine respiratory disease (BRD) poses a persistent challenge in the beef cattle industry, impacting both animal health and economic aspects. Several risk factors make an animal susceptible to BRD, including bacteria such as Mannheimia haemolytica, Pasteurella multocida, Histophilus somni, and Mycoplasma bovis. Despite efforts to characterize and quantify these bacteria in the nasal cavity for disease diagnosis, more research is needed to understand if there is a pathobiont abundance threshold for clinical signs of respiratory disease, and if the results are similar across feedlots. This study aims to compare the nasal microbiome community diversity and composition, along with the abundance of four bacterial pathogens and associated serotypes, in apparently healthy and BRD-affected beef cattle. Nasal swabs were collected from four beef feedlots across the US, covering the years 2019 to 2022. The study included post-weaned beef cattle with diverse housing conditions.
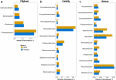
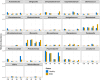
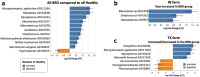
Results: Quantification of BRD-associated pathogens effectively distinguished BRD-affected from apparently healthy beef cattle, surpassing the efficacy of 16S rRNA gene sequencing of the nasal microbiome community. Specifically, H. somni, M. bovis, and M. haemolytica had higher abundance in the BRD-affected group. Utilizing the abundance of these pathobionts and analyzing their combined abundance with machine learning models resulted in an accuracy of approximately 63% for sample classification into disease status. Moreover, there were no significant differences in nasal microbiome diversity (alpha and beta) between BRD-affected and apparently healthy cattle; instead, differences were detected between feedlots.
Conclusions: Notably, this study sheds light on the beef cattle nasal microbiome community composition, revealing specific differences between BRD-affected and apparently healthy cattle. Pathobiont abundance was increased in some, but not all farms. Nonetheless, more research is needed to determine if these differences are consistent across other studies. Additionally, future research should consider bacterial-viral interactions in the beef nasal metagenome.
Keywords: 16S rRNA gene; BRD-pathobionts; Beef cattle; Bovine respiratory disease; qPCR.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: All procedures involving animal use were approved by the Purdue University Animal Care and Use Committee (protocol #1906001911). Consent for publication: All authors provide consent to publish. Competing interests: The authors declare no competing interests.
Figures



References
-
- Casella E, Cantor MC, Silvestri S, Renaud DL, Costa JHC. Cost-aware inference of bovine respiratory disease in calves using precision livestock technology. Proc – 18th Annual Int Conf Distrib Comput Sens Syst DCOSS 2022. 2022;109–116. 10.1109/DCOSS54816.2022.00031.
-
- Snowder GD, Van Vleck LD, Cundiff LV, Bennett GL. Bovine respiratory disease in feedlot cattle: environmental, genetic, and economic factors. J Anim Sci. 2006;84:1999–2008. 10.2527/jas.2006-046. - PubMed
-
- USDA. Feedlot 2011. March; 2013. p. 154.
-
- Edward AJ. Respiratory disease in feedlot cattle in the central USA. Bov Pr. 1996;30:5–7.
-
- Ferraro S, Fecteau G, Dubuc J, Francoz D, Rousseau M, Roy JP, Buczinski S. Scoping review on clinical definition of bovine respiratory disease complex and related clinical signs in dairy cows. J Dairy Sci. 2021;104:7095–108. 10.3168/jds.2020-19471. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
