Regulation of acetyl-CoA biosynthesis via an intertwined acetyl-CoA synthetase/acetyltransferase complex
- PMID: 40089509
- PMCID: PMC11910552
- DOI: 10.1038/s41467-025-57842-2
Regulation of acetyl-CoA biosynthesis via an intertwined acetyl-CoA synthetase/acetyltransferase complex
Abstract
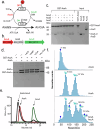
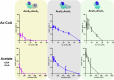
Acetyl-CoA synthetase (Acs) generates acetyl-coenzyme A (Ac-CoA) but its excessive activity can deplete ATP and lead to a growth arrest. To prevent this, Acs is regulated through Ac-CoA-dependent feedback inhibition executed by Ac-CoA-dependent acetyltransferases such as AcuA in Bacillus subtilis. AcuA acetylates the catalytic lysine of AcsA turning the synthetase inactive. Here, we report that AcuA and AcsA form a tightly intertwined complex - the C-terminal domain binds to acetyltransferase domain of AcuA, while the C-terminus of AcuA occupies the CoA-binding site in the N-terminal domain of AcsA. Formation of the complex reduces AcsA activity in addition to the well-established acetylation of the catalytic lysine 549 in AcsA which we show can disrupt the complex. Thus, different modes of regulation accomplished through AcuA adjust AcsA activity to the concentrations of the different substrates of the reaction. In summary, our study provides detailed mechanistic insights into the regulatory framework underlying acetyl-CoA biosynthesis from acetate.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

