Ubiquinol-mediated suppression of mitochondria-associated ferroptosis is a targetable function of lactate dehydrogenase B in cancer
- PMID: 40090955
- PMCID: PMC11911438
- DOI: 10.1038/s41467-025-57906-3
Ubiquinol-mediated suppression of mitochondria-associated ferroptosis is a targetable function of lactate dehydrogenase B in cancer
Abstract
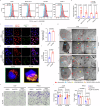
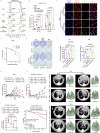
Lactate dehydrogenase B (LDHB) fuels oxidative cancer cell metabolism by converting lactate to pyruvate. This study uncovers LDHB's role in countering mitochondria-associated ferroptosis independently of lactate's function as a carbon source. LDHB silencing alters mitochondrial morphology, causes lipid peroxidation, and reduces cancer cell viability, which is potentiated by the ferroptosis inducer RSL3. Unlike LDHA, LDHB acts in parallel with glutathione peroxidase 4 (GPX4) and dihydroorotate dehydrogenase (DHODH) to suppress mitochondria-associated ferroptosis by decreasing the ubiquinone (coenzyme Q, CoQ) to ubiquinol (CoQH2) ratio. Indeed, supplementation with mitoCoQH2 (mitochondria-targeted analogue of CoQH2) suppresses mitochondrial lipid peroxidation and cell death after combined LDHB silencing and RSL3 treatment, consistent with the presence of LDHB in the cell fraction containing the mitochondrial inner membrane. Addressing the underlying molecular mechanism, an in vitro NADH consumption assay with purified human LDHB reveals that LDHB catalyzes the transfer of reducing equivalents from NADH to CoQ and that the efficiency of this reaction increases by the addition of lactate. Finally, radiation therapy induces mitochondrial lipid peroxidation and reduces tumor growth, which is further enhanced when combined with LDHB silencing. Thus, LDHB-mediated lactate oxidation drives the CoQ-dependent suppression of mitochondria-associated ferroptosis, a promising target for combination therapies.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
-
- Warburg, O. & Minami, S. Versuche an Überlebendem Carcinom-gewebe. Klinische Wochenschr.2, 776–777 (1923).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

