Conditional Overexpression of Serpine2 Promotes Hair Cell Regeneration from Lgr5+ Progenitors in the Neonatal Mouse Cochlea
- PMID: 40091489
- PMCID: PMC12079390
- DOI: 10.1002/advs.202412653
Conditional Overexpression of Serpine2 Promotes Hair Cell Regeneration from Lgr5+ Progenitors in the Neonatal Mouse Cochlea
Abstract
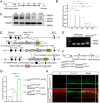
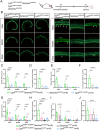
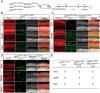
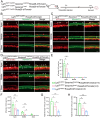
Neonatal cochlear Lgr5+ progenitors retain limited hair cells (HCs) regenerative capacity, but the regulatory network remains incompletely defined. Serpin family E member 2 (Serpine2) is shown to participate in regulating proliferation and differentiation of cochlear Lgr5+ progenitors in the previous in vitro study. Here, the expression pattern and in vivo roles of Serpine2 in HC regeneration are explored by transgenic mice. It is found that Serpine2 is expressed in the mouse cochlea after birth with a downward trend as the mice age. In addition, Serpine2 conditional overexpression in vivo in Lgr5+ progenitors of neonatal mice cochlea results in an increased number of ectopic HCs in a dose-dependent manner. Serpine2 knockdown ex vivo and in vivo can inhibit HC regeneration. EdU assay and lineage tracing assay demonstrate these ectopic HCs likely originate from Lgr5+ progenitors through direct transdifferentiation rather than through mitotic regeneration. Moreover, single-nucleus RNA sequencing analysis and mRNA level validation reveal that conditionally overexpressed Serpine2 likely induces HC regeneration via inhibiting sonic hedgehog (SHH) signal pathway and inducing Atoh1 and Pou4f3 transcription factor. In brief, these data indicate that Serpine2 plays a pivotal role in HC regeneration from Lgr5+ progenitors in the neonatal mouse cochlea, and this suggests a new avenue for future research into HC regeneration.
Keywords: Lgr5+ progenitors; Serpine2; hair cells; regeneration.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Conditional Overexpression of Net1 Enhances the Trans-Differentiation of Lgr5+ Progenitors into Hair Cells in the Neonatal Mouse Cochlea.Cell Prolif. 2025 Apr;58(4):e13787. doi: 10.1111/cpr.13787. Epub 2024 Dec 15. Cell Prolif. 2025. PMID: 39675772 Free PMC article.
-
Knockdown of Foxg1 in supporting cells increases the trans-differentiation of supporting cells into hair cells in the neonatal mouse cochlea.Cell Mol Life Sci. 2020 Apr;77(7):1401-1419. doi: 10.1007/s00018-019-03291-2. Epub 2019 Sep 4. Cell Mol Life Sci. 2020. PMID: 31485717 Free PMC article.
-
Characterization of Lgr5+ progenitor cell transcriptomes in the apical and basal turns of the mouse cochlea.Oncotarget. 2016 Jul 5;7(27):41123-41141. doi: 10.18632/oncotarget.8636. Oncotarget. 2016. PMID: 27070092 Free PMC article.
-
Postnatal development, maturation and aging in the mouse cochlea and their effects on hair cell regeneration.Hear Res. 2013 Mar;297:68-83. doi: 10.1016/j.heares.2012.11.009. Epub 2012 Nov 16. Hear Res. 2013. PMID: 23164734 Free PMC article. Review.
-
Understanding the differentiation and epigenetics of cochlear sensory progenitors in pursuit of regeneration.Curr Opin Otolaryngol Head Neck Surg. 2021 Oct 1;29(5):366-372. doi: 10.1097/MOO.0000000000000741. Curr Opin Otolaryngol Head Neck Surg. 2021. PMID: 34374667 Free PMC article. Review.
References
-
- a) Shu Y., Li W., Huang M., Quan Y. Z., Scheffer D., Tian C., Tao Y., Liu X., Hochedlinger K., Indzhykulian A. A., Wang Z., Li H., Chen Z. Y., Nat. Commun. 2019, 10, 5530; - PMC - PubMed
- b) Wagner E. L., Shin J. B., Trends Neurosci. 2019, 42, 414; - PMC - PubMed
- c) Sato M. P., Benkafadar N., Heller S., Cell Rep. 2024, 43, 113822. - PMC - PubMed
-
- a) García‐Añoveros J., Clancy J. C., Foo C. Z., García‐Gómez I., Zhou Y., Homma K., Cheatham M. A., Duggan A., Nature 2022, 605, 298; - PMC - PubMed
- b) Roccio M., Edge A. S. B., Development 2019, 146, dev177188; - PMC - PubMed
- c) Tan F., Chu C., Qi J., Li W., You D., Li K., Chen X., Zhao W., Cheng C., Liu X., Qiao Y., Su B., He S., Zhong C., Li H., Chai R., Zhong G., Nat. Commun. 2019, 10, 3733; - PMC - PubMed
- d) Li X. J., Morgan C., Goff L. A., Doetzlhofer A., Sci. Adv. 2022, 8, eabj7651. - PMC - PubMed
-
- Barker N., van Es J. H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P. J., Clevers H., Nature 2007, 449, 1003. - PubMed
-
- Barker N., Huch M., Kujala P., van de Wetering M., Snippert H. J., van Es J. H., Sato T., Stange D. E., Begthel H., van den Born M., Danenberg E., van den Brink S., Korving J., Abo A., Peters P. J., Wright N., Poulsom R., Clevers H., Cell Stem Cell 2010, 6, 25. - PubMed
MeSH terms
Substances
Grants and funding
- 2023YFA1801804/National Key R&D Program of China
- 2022YFA0807000/National Key R&D Program of China
- 2021YFA1101300/National Key R&D Program of China
- 2021YFA1101800/National Key R&D Program of China
- 2020YFA0112503/National Key R&D Program of China
- 82171149/National Natural Science Foundation of China
- 82371166/National Natural Science Foundation of China
- 81970892/National Natural Science Foundation of China
- 82030029/National Natural Science Foundation of China
- 81970882/National Natural Science Foundation of China
- 92149304/National Natural Science Foundation of China
- 2024A1515010548/Guangdong Basic and Applied Basic Research Foundation
- JCYJ20230807114700001/Shenzhen Science and Technology Program
- JCYJ20190814093401920/Shenzhen Science and Technology Program
- JCYJ20210324125608022/Shenzhen Science and Technology Program
- XDA16010303/Strategic Priority Research Program of the Chinese Academy of Science
- 2021YFS0371/Science and Technology Department of Sichuan Province
- SKLGE-2104/Open Research Fund of the State Key Laboratory of Genetic Engineering, Fudan University
- BK20233002/Jiangsu Provincial Scientific Research Center of Applied Mathematics
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
