DNA methylation confers a cerebellum-specific identity in non-human primates
- PMID: 40091535
- PMCID: PMC12000133
- DOI: 10.24272/j.issn.2095-8137.2024.325
DNA methylation confers a cerebellum-specific identity in non-human primates
Abstract
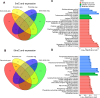
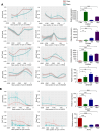
Selective regulation of gene expression across distinct brain regions is crucial for establishing and maintaining subdivision identities. DNA methylation, a key regulator of gene transcription, modulates transcriptional activity through the conversion of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC). While DNA methylation is hypothesized to play an essential role in shaping brain identity by influencing gene expression patterns, its direct contribution, especially in primates, remains largely unexplored. This study examined DNA methylation landscapes and transcriptional profiles across four brain regions, including the cortex, cerebellum, striatum, and hippocampus, using samples from 12 rhesus monkeys. The cerebellum exhibited distinct epigenetic and transcriptional signatures, with differentially methylated regions (DMRs) significantly enriched in metabolic pathways. Notably, genes harboring clustered differentially hydroxymethylated regions (DhMRs) overlapped with those implicated in schizophrenia. Moreover, 5mC located 1 kb upstream of the ATG start codon was correlated with gene expression and exhibited region-specific associations with 5hmC. These findings provide insights into the coordinated regulation of cerebellum-specific 5mC and 5hmC , highlighting their potential roles in defining cerebellar identity and contributing to neuropsychiatric diseases.
基因的选择性表达对于建立和维持不同大脑区域的身份至关重要。DNA甲基化是重要的基因转录调节因子,包含5-甲基胞嘧啶 (5mC) 到5-羟甲基胞嘧啶 (5hmC) 的转化。研究认为DNA甲基化通过影响基因表达模式在调节大脑身份方面发挥重要作用。然而,DNA甲基化与大脑特征的直接关联,尤其是在灵长类动物的大脑中仍不清楚。该研究使用12只猕猴,对四个大脑区域(皮质、小脑、纹状体和海马)的全基因组DNA甲基化和基因转录模式进行了全面分析。我们的研究揭示了小脑中5mC、5hmC和基因表达的独特模式。值得注意的是,猕猴小脑中的差异甲基化区域(DMRs)在代谢途径中丰富。有趣的是,一群差异羟甲基化(DhMRs)富集的基因与精神分裂症相关基因重叠。此外,我们发现位于ATG起始密码子上游1 kilobase的5mC主导基因表达,5hmC以两种方式参与调控过程。这些发现揭示了小脑特异性5mC和5hmC对基因表达的协同调节,为小脑特性和相关疾病的研究提供数据积累。.
Keywords: 5-Hydroxymethylcytosine; 5-Methylcytosine; Cerebellum; Gene expression; Non-human primates; Tissue identity.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





Similar articles
-
Distal regulatory elements identified by methylation and hydroxymethylation haplotype blocks from mouse brain.Epigenetics Chromatin. 2018 Dec 29;11(1):75. doi: 10.1186/s13072-018-0248-3. Epigenetics Chromatin. 2018. PMID: 30594220 Free PMC article.
-
Genome-wide characterization of dynamic DNA 5-hydroxymethylcytosine and TET2-related DNA demethylation during breast tumorigenesis.Clin Epigenetics. 2024 Sep 11;16(1):125. doi: 10.1186/s13148-024-01726-7. Clin Epigenetics. 2024. PMID: 39261937 Free PMC article.
-
Association of 5-hydroxymethylation and 5-methylation of DNA cytosine with tissue-specific gene expression.Epigenetics. 2017 Feb;12(2):123-138. doi: 10.1080/15592294.2016.1265713. Epub 2016 Dec 2. Epigenetics. 2017. PMID: 27911668 Free PMC article.
-
Distinctive Patterns of 5-Methylcytosine and 5-Hydroxymethylcytosine in Schizophrenia.Int J Mol Sci. 2024 Jan 4;25(1):636. doi: 10.3390/ijms25010636. Int J Mol Sci. 2024. PMID: 38203806 Free PMC article. Review.
-
The elusive role of 5'-hydroxymethylcytosine.Epigenomics. 2016 Nov;8(11):1539-1551. doi: 10.2217/epi-2016-0076. Epub 2016 Oct 13. Epigenomics. 2016. PMID: 27733056 Review.
References
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
