Metabolic insights of lactic acid bacteria in reducing off-flavors and antinutrients in plant-based fermented dairy alternatives
- PMID: 40091739
- PMCID: PMC11911983
- DOI: 10.1111/1541-4337.70134
Metabolic insights of lactic acid bacteria in reducing off-flavors and antinutrients in plant-based fermented dairy alternatives
Abstract
Multiple sensorial, technological, and nutritional challenges must be overcome when developing plant-based fermented dairy alternatives (PBFDA) to mimic their dairy counterparts. The elimination of plant-derived off-flavors (green, earthy, bitter, astringent) and the degradation of antinutrients are crucial quality factors highlighted by the industry for their effect on consumer acceptance. The adaptation of plant-derived lactic acid bacteria (LAB) species into plant niches is relevant when developing starter cultures for PBFDA products due to their evolutionary acquired ability to degrade plant-based undesirable compounds (off-flavors and antinutrients). Some plant-isolated species, such as Lactiplantibacillus plantarum and Limosilactobacillus fermentum, have been associated with the degradation of phytates, phenolic compounds, oxalates, and raffinose-family oligosaccharides (RFOs), whereas some animal-isolated species, such as Lactobacillus acidophilus strains, can metabolize phytates, RFOs, saponins, phenolic compounds, and oxalates. Some proteolytic LAB strains, such as Lacticaseibacillus paracasei and Lacticaseibacillus rhamnosus, have been characterized to degrade phytates, protease inhibitors, and oxalates. Other species have also been described regarding their abilities to biotransform phytic acid, RFOs, saponins, phenolic compounds, protease inhibitors, oxalates, and volatile off-flavor compounds (hexanal, nonanal, pentanal, and benzaldehyde). In addition, we performed a blast analysis considering antinutrient metabolic genes (42 genes) to up to 5 strains of all qualified presumption of safety-listed LAB species (55 species, 240 strains), finding out potential genotypical capabilities of LAB species that have not conventionally been used as starter cultures such as Lactiplantibacillus pentosus, Lactiplantibacillus paraplantarum, and Lactobacillus diolivorans for plant-based fermentations. This review provides a detailed understanding of genes and enzymes from LAB that target specific compounds in plant-based materials for plant-based fermented food applications.
Keywords: antinutrients; fermentation; lactic acid bacteria; off‐flavors; plant‐based alternatives.
© 2025 The Author(s). Comprehensive Reviews in Food Science and Food Safety published by Wiley Periodicals LLC on behalf of Institute of Food Technologists.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

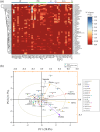
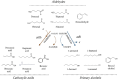
References
-
- Azcarate‐Peril, M. A. , Bruno‐Bárcena, J. M. , Hassan, H. M. , & Klaenhammer, T. R. (2006). Transcriptional and functional analysis of oxalyl‐coenzyme A (CoA) decarboxylase and formyl‐CoA transferase genes from Lactobacillus acidophilus . Applied and Environmental Microbiology, 72(3), 1891–1899. 10.1128/AEM.72.3.1891-1899.2006 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

