Unraveling the role of proteins in dementia: insights from two UK cohorts with causal evidence
- PMID: 40092369
- PMCID: PMC11906402
- DOI: 10.1093/braincomms/fcaf097
Unraveling the role of proteins in dementia: insights from two UK cohorts with causal evidence
Abstract
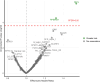
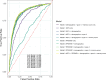
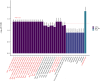
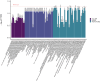
Population-based proteomics offers a groundbreaking avenue to predict future disease risks, enhance our understanding of disease mechanisms, and discover novel therapeutic targets and biomarkers. The role of plasma proteins in dementia, however, requires further exploration. This study investigated 276 protein-dementia associations in 229 incident all-cause dementia, 89 Alzheimer's disease, and 41 vascular dementia among 3249 participants (55% women, 97.2% white ethnicity) from the English Longitudinal Study of Ageing (ELSA) over a median 9.8-year follow-up. We used Cox proportional hazard regression for the analysis. Receiver operating characteristic analyses were conducted to assess the precision of the identified proteins from the fully adjusted Cox regression models in predicting incident all-cause dementia, both individually and in combination with demographic predictors, APOE genotype, and memory score, to estimate the area under the curve. Additionally, the eXtreme Gradient Boosting machine learning algorithm was used to identify the most important features predictive of future all-cause dementia onset. These associations were then validated in 1506 incident all-cause dementia, 732 Alzheimer's disease, 281 vascular dementia, and 111 frontotemporal dementia cases among 52 745 individuals (53.9% women, 93.3% White ethnicity) from the UK Biobank over a median 13.7-year follow-up. Two-sample bi-directional Mendelian randomization and drug target Mendelian randomization were further employed to determine the causal direction between protein concentration and dementia. NEFL (hazard ratio [HR] [95% confidence intervals (CIs)]: 1.54 [1.29, 1.84]) and RPS6KB1 (HR [95% CI]: 1.33 [1.16, 1.52]) were robustly associated with incident all-cause dementia; MMP12 (HR [95% CI]: 2.06 [1.41, 2.99]) was associated with vascular dementia in ELSA, after correcting for multiple testing. Additional markers EDA2R and KIM1 were identified from subgroup and sensitivity analyses. Combining NEFL and RPS6KB1 with other predictors yielded high predictive accuracy (area under the curve = 0.871) for incident all-cause dementia. The eXtreme Gradient Boosting machine learning algorithm also identified RPS6KB1, NEFL, and KIM1 as the most important protein features for predicting future all-cause dementia. Sex difference was evident for the association between RPS6KB1 and all-cause dementia, with stronger association in men (P for interaction = 0.037). Replication in the UK Biobank confirmed the associations between the identified proteins and various dementia subtypes. The results from Mendelian randomization in the reverse direction indicated that several proteins serve as early markers for dementia, rather than being direct causes of the disease. These findings provide insights into putative mechanisms for dementia. Future studies are needed to validate the findings on RPS6KB1 in relation to dementia risk.
Keywords: ELSA; Mendelian randomization; UK biobank; dementia; proteomics.
© The Author(s) 2025. Published by Oxford University Press on behalf of the Guarantors of Brain.
Conflict of interest statement
Olink had no part in designing the study or analyzing the data. No conflicts of interest to be declared from any of the authors.
Figures



Update of
-
Unraveling the role of plasma proteins in dementia: insights from two cohort studies in the UK, with causal evidence from Mendelian randomization.medRxiv [Preprint]. 2024 Jun 4:2024.06.04.24308415. doi: 10.1101/2024.06.04.24308415. medRxiv. 2024. Update in: Brain Commun. 2025 Mar 03;7(2):fcaf097. doi: 10.1093/braincomms/fcaf097. PMID: 38883777 Free PMC article. Updated. Preprint.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
