Rational design of gold nanoparticle-based chemosensors for detection of the tumor marker 3-methoxytyramine
- PMID: 40092597
- PMCID: PMC11908650
- DOI: 10.1039/d4sc08758e
Rational design of gold nanoparticle-based chemosensors for detection of the tumor marker 3-methoxytyramine
Abstract
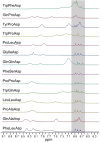
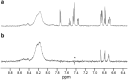
In this study, we combined computational modeling, simulations, and experiments to design gold nanoparticle-based receptors specifically tailored for the NMR detection of 3-methoxytyramine (3-MT), a prognostic marker for asymptomatic neuroblastoma. We used short steered MD simulations to rank a library of 100 newly functionalized, tripeptide-coated AuNPs for their ability to recognize 3-MT. Validation of the computational analysis was performed on a subset of synthesized tripeptide-coated nanoparticles, showing a strong correlation between the predicted and experimental affinities. Eventually, we tested the sensing performance using nanoparticle-assisted NMR chemosensing, a technique which relies on magnetization transfer within a nanoparticle-host/analyte-guest complex to isolate the sole NMR signals of the analyte. This approach led to the identification of novel chemosensors that exhibited better performance compared to existing ones, lowering the limit of detection below 25 μM and demonstrating the utility of this integrated strategy.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures




References
-
- Eersels K. Lieberzeit P. Wagner P. ACS Sens. 2016;1:1171–1187. doi: 10.1021/acssensors.6b00572. - DOI
LinkOut - more resources
Full Text Sources

