This is a preprint.
P4-ATPase control over phosphoinositide membrane asymmetry and neomycin resistance
- PMID: 40093091
- PMCID: PMC11908233
- DOI: 10.1101/2025.03.03.641220
P4-ATPase control over phosphoinositide membrane asymmetry and neomycin resistance
Update in
-
P4-ATPases control phosphoinositide membrane asymmetry and neomycin resistance.Nat Cell Biol. 2025 Jul;27(7):1114-1124. doi: 10.1038/s41556-025-01692-z. Epub 2025 Jul 11. Nat Cell Biol. 2025. PMID: 40646185 Free PMC article.
Abstract
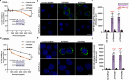
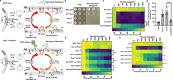
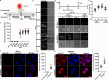
Neomycin, an aminoglycoside antibiotic, has robust antibacterial properties, yet its clinical utility is curtailed by its nephrotoxicity and ototoxicity. The mechanism by which the polycationic neomycin enters specific eukaryotic cell types remains poorly understood. In budding yeast, NEO1 is required for neomycin resistance and encodes a phospholipid flippase that establishes membrane asymmetry. Here, we show that mutations altering Neo1 substrate recognition cause neomycin hypersensitivity by exposing phosphatidylinositol-4-phosphate (PI4P) in the plasma membrane extracellular leaflet. Human cells also expose extracellular PI4P upon knockdown of ATP9A, a Neo1 ortholog and ATP9A expression level correlates to neomycin sensitivity. In yeast, the extracellular PI4P is initially produced in the cytosolic leaflet of the plasma membrane and then delivered by Osh6-dependent nonvesicular transport to the endoplasmic reticulum (ER). Here, a portion of PI4P escapes degradation by the Sac1 phosphatase by entering the ER lumenal leaflet. COPII vesicles transport lumenal PI4P to the Golgi where Neo1 flips this substrate back to the cytosolic leaflet. Cryo-EM reveals that PI4P binds Neo1 within the substrate translocation pathway. Loss of Neo1 activity in the Golgi allows secretion of extracellular PI4P, which serves as a neomycin receptor and facilitates its endocytic uptake. These findings unveil novel mechanisms of aminoglycoside sensitivity and phosphoinositide homeostasis, with important implications for signaling by extracellular phosphoinositides.
Conflict of interest statement
Conflict of interest: The authors declare no competing interests.
Figures



References
-
- Waksman S. A. & Lechevalier H. A. Neomycin, a New Antibiotic Active against Streptomycin-Resistant Bacteria, including Tuberculosis Organisms. Science 109, 305–307 (1949). - PubMed
-
- ROBSON J. M. & SULLIVAN F. M. Antituberculosis drugs. Pharmacol. Rev. 15, 169–223 (1963). - PubMed
-
- Davies J. & Davis B. D. Misreading of ribonucleic acid code words induced by aminoglycoside antibiotics. The effect of drug concentration. J. Biol. Chem. 243, 3312–3316 (1968). - PubMed
-
- Davies J., Gorini L. & Davis B. D. Misreading of RNA codewords induced by aminoglycoside antibiotics. Mol. Pharmacol. 1, 93–106 (1965). - PubMed
-
- ANDRE T. Studies on the distribution of tritium-labelled dihydrostreptomycin and tetracycline in the body. Acta Radiol. Suppl. 1–89 (1956). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
