This is a preprint.
Independent signaling pathways provide a fail-safe mechanism to prevent tumorigenesis
- PMID: 40093137
- PMCID: PMC11908167
- DOI: 10.1101/2025.02.28.640798
Independent signaling pathways provide a fail-safe mechanism to prevent tumorigenesis
Abstract
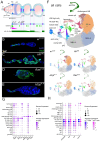
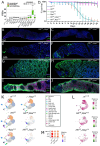
Controlled signaling activity is vital for normal tissue homeostasis and oncogenic signaling activation facilitates tumorigenesis. Here we use single-cell transcriptomics to investigate the effects of pro-proliferative signaling on epithelial homeostasis using the Drosophila follicle cell lineage. Notably, EGFR-Ras overactivation induces cell cycle defects by activating the transcription factors Pointed and E2f1 and impedes differentiation. Hh signaling simultaneously promotes an undifferentiated state and induces differentiation via activation of EMT-associated transcription factors zfh1 and Mef2. As a result, overactivation of Hh signaling generates a transcriptional hybrid state comparable to epithelial-mesenchymal-transition. Co-overactivation of Hh signaling with EGFR-Ras signaling blocks differentiation and induces key characteristics of tumor cells including a loss of tissue architecture caused by reduced expression of cell adhesion molecules, sustained proliferation and an evasion of cell cycle checkpoints. These findings provide new insight into how non-interacting signaling pathways converge at the transcriptional level to prevent malignant cell behavior.
Figures




References
-
- Chacón-Martínez C.A., Koester J., and Wickström S.A. (2018). Signaling in the stem cell niche: regulating cell fate, function and plasticity. Development 145, dev165399. - PubMed
-
- Bahar M.E., Kim H.J., and Kim D. (2023). Targeting the RAS/RAF/MAPK pathway for cancer therapy: from mechanism to clinical studies. Signal Transduct. Target. Ther. 8. 10.1038/s41392-023-01705-z. - DOI
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
