This is a preprint.
GA4GH Phenopacket-Driven Characterization of Genotype-Phenotype Correlations in Mendelian Disorders
- PMID: 40093222
- PMCID: PMC11908317
- DOI: 10.1101/2025.03.05.25323315
GA4GH Phenopacket-Driven Characterization of Genotype-Phenotype Correlations in Mendelian Disorders
Abstract
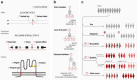
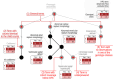
Comprehensively characterizing genotype-phenotype correlations (GPCs) in Mendelian disease would create new opportunities for improving clinical management and understanding disease biology. However, heterogeneous approaches to data sharing, reuse, and analysis have hindered progress in the field. We developed Genotype Phenotype Evaluation of Statistical Association (GPSEA), a software package that leverages the Global Alliance for Genomics and Health (GA4GH) Phenopacket Schema to represent case-level clinical and genetic data about individuals. GPSEA applies an independent filtering strategy to boost statistical power to detect categorical GPCs represented by Human Phenotype Ontology terms. GPSEA additionally enables visualization and analysis of continuous phenotypes, clinical severity scores, and survival data such as age of onset of disease or clinical manifestations. We applied GPSEA to 85 cohorts with 6613 previously published individuals with variants in one of 80 genes associated with 122 Mendelian diseases and identified 225 significant GPCs, with 48 cohorts having at least one statistically significant GPC. These results highlight the power of standardized representations of clinical data for scalable discovery of GPCs in Mendelian disease.
Conflict of interest statement
Competing interests The authors declare no competing interests
Figures


References
-
- Ries M. & Gal A. Genotype–phenotype correlation in Fabry disease. in Fabry Disease: Perspectives from 5 Years of FOS (eds. Mehta A., Beck M. & Sunder-Plassmann G.) (Oxford PharmaGenesis, Oxford, 2006). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
