PFMG2025-integrating genomic medicine into the national healthcare system in France
- PMID: 40093400
- PMCID: PMC11910791
- DOI: 10.1016/j.lanepe.2024.101183
PFMG2025-integrating genomic medicine into the national healthcare system in France
Abstract
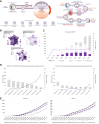
Integrating genomic medicine into healthcare systems is a health policy challenge that requires continuously transferring scientific advances into clinics and ensuring equal access for patients. France was one of the first countries to integrate genome sequencing into clinical practice at a nationwide level, with the ambition to provide more accurate diagnostics and personalized treatments. Since 2016, the French government has invested €239M in the 2025 French Genomic Medicine Initiative (PFMG2025) which has so far focused on patients with rare diseases (RD), cancer genetic predisposition (CGP) and cancers. PFMG2025 has addressed numerous challenges to set up an operational organizational framework. As of December the 31st 2023, 12,737 results were returned to prescribers for RD/CGP patients (median delivery time: 202 days, diagnostic yield: 30.6%) and 3109 for cancer patients (median delivery time: 45 days). PFMG2025's future priorities encompass ensuring economic sustainability, strengthening links with research, empowering patients and practitioners, and fostering collaborations with European partners.
Funding: As of December the 31st 2023, €239M have been invested by the French government.
Keywords: Cancer predisposition; Cancers; French genomic medicine initiative; Genome sequencing; Genomic medicine; PFMG2025; Rare diseases.
© 2024 The Authors.
Conflict of interest statement
PFMG2025 leadership declare no conflict of interest. J. Y Blay has relationships with the INCa, the EU commission and the French National Research Agency (ANR) and has received research grant for the clinical trial Profiler 2 (not related) from the Roche company. P. Saintigny and has received grant and equipment, materials, drugs, medical writing, gifts or other services from the Roche, Roche Molecular Diagnostics, Astrazeneca, Novartis, Bristol Myer Squibb, Illumina, HTG Molecular Diagnostics, Inivita, Archer, Omicure, Smartcatch and ADMIR companies, as well as the BMS Foundation. P. Laurent-Puig is the President of the Cancéropole Ile-de-France, has stock options in the MethysDX company and has received consulting fees from the Biocartis, Amgen, Pierre Fabre and Servier companies, as well as the BMS Foundation.
Figures


References
-
- Stenzinger A., Edsjo A., Ploeger C., et al. Trailblazing precision medicine in Europe: a joint view by genomic medicine Sweden and the centers for personalized medicine, ZPM, in Germany. Semin Cancer Biol. 2022;84:242–254. - PubMed
-
- Stenzinger A., Moltzen E.K., Winkler E., et al. Implementation of precision medicine in healthcare-A European perspective. J Intern Med. 2023;294:437–454. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

