Development of p300-targeting degraders with enhanced selectivity and onset of degradation
- PMID: 40093518
- PMCID: PMC11905989
- DOI: 10.1039/d4md00969j
Development of p300-targeting degraders with enhanced selectivity and onset of degradation
Abstract
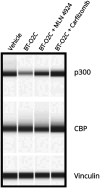
p300 and CBP are paralogous epigenetic regulators that are considered promising therapeutic targets for cancer treatment. Small molecule p300/CBP inhibitors have so far been unable to differentiate between these closely related proteins, yet selectivity is desirable in order to probe their distinct cellular functions. Additionally, in multiple cancers, loss-of-function CREBBP mutations set up a paralog dependent synthetic lethality with p300, that could be exploited with a selective therapeutic agent. To address this, we developed p300-targeting heterobifunctional degraders that recruit p300 through its HAT domain using the potent spiro-hydantoin-based inhibitor, iP300w. Lead degrader, BT-O2C, demonstrates improved selectivity and a faster onset of action compared to a recently disclosed A 485-based degrader in HAP1 cells and is cytotoxic in CIC::DUX4 sarcoma (CDS) cell lines (IC50 = 152-221 nM), significantly reducing expression of CDS target genes (ETV1, ETV4, ETV5). Taken together, our results demonstrate that BT-O2C represents a useful tool degrader for further exploration of p300 degradation as a therapeutic strategy.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There is no conflict of interest to declare.
Figures






References
-
- Lasko L. M. Jakob C. G. Edalji R. P. Qiu W. Montgomery D. Digiammarino E. L. Hansen T. M. Risi R. M. Frey R. Manaves V. Shaw B. Algire M. Hessler P. Lam L. T. Uziel T. Faivre E. Ferguson D. Buchanan F. G. Martin R. L. Torrent M. Chiang G. G. Karukurichi K. Langston J. W. Weinert B. T. Choudhary C. de Vries P. Kluge A. F. Patane M. A. Drie J. H. V. Wang C. McElligott D. Kesicki E. Marmorstein R. Sun C. Cole P. A. Rosenberg S. H. Michaelides M. R. Lai A. Bromberg K. D. Discovery of a Selective Catalytic P300/CBP Inhibitor That Targets Lineage-Specific Tumours. Nature. 2017;550(7674):128–132. doi: 10.1038/nature24028. https://dx.doi.org/10.1038/nature24028 - DOI - DOI - PMC - PubMed
-
- Ogryzko V. V. Schiltz R. L. Russanova V. Howard B. H. Nakatani Y. The Transcriptional Coactivators P300 and CBP Are Histone Acetyltransferases. Cell. 1996;87(5):953–959. doi: 10.1016/S0092-8674(00)82001-2. https://dx.doi.org/10.1016/S0092-8674(00)82001-2 - DOI - DOI - PubMed
-
- Bannister A. J. Kouzarides T. The CBP Co-Activator Is a Histone Acetyltransferase. Nature. 1996;384(6610):641–643. doi: 10.1038/384641a0. https://dx.doi.org/10.1038/384641a0 - DOI - DOI - PubMed
-
- Hnisz D. Abraham B. J. Lee T. I. Lau A. Saint-André V. Sigova A. A. Hoke H. A. Young R. A. Super-Enhancers in the Control of Cell Identity and Disease. Cell. 2013;155(4):934–947. doi: 10.1016/j.cell.2013.09.053. https://dx.doi.org/10.1016/j.cell.2013.09.053 - DOI - DOI - PMC - PubMed
-
- Dancy B. M. Cole P. A. Protein Lysine Acetylation by P300/CBP. Chem. Rev. 2015;115(6):2419–2452. doi: 10.1021/cr500452k. https://dx.doi.org/10.1021/cr500452k - DOI - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

