LCORL and STC2 Variants Increase Body Size and Growth Rate in Cattle and Other Animals
- PMID: 40094447
- PMCID: PMC12448305
- DOI: 10.1093/gpbjnl/qzaf025
LCORL and STC2 Variants Increase Body Size and Growth Rate in Cattle and Other Animals
Abstract
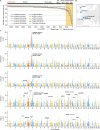
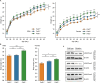
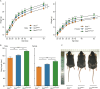
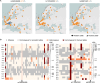
Natural variants can significantly improve growth traits in livestock and serve as safe targets for gene editing, thus being applied in animal molecular design breeding. However, such safe and large-effect mutations are severely lacking. Using ancestral recombination graphs, we investigated recent selection signatures in beef cattle breeds, pinpointing sweep-driving variants in the LCORL and STC2 loci with notable effects on body size and growth rate. The ACT-to-A frameshift mutation in LCORL occurs mainly in central-European cattle, and stimulates growth. Remarkably, convergent truncating mutations were also found in commercial breeds of sheep, goats, pigs, horses, dogs, rabbits, and chickens. In the STC2 gene, we identified a missense mutation (A60P) located within the conserved region across vertebrates. We validated the two natural mutations in gene-edited mouse models, where both variants in homozygous carriers significantly increase the average weight by 11%. Our findings provide insights into a seemingly recurring gene target of body size enhancing truncating mutations across domesticated species, and offer valuable targets for gene editing-based breeding in animals.
Keywords: LCORL; STC2; Ancestral recombination graph; Common improving gene; Convergent selection.
© The Author(s) 2025. Published by Oxford University Press and Science Press on behalf of the Beijing Institute of Genomics, Chinese Academy of Sciences / China National Center for Bioinformation and Genetics Society of China.
Conflict of interest statement
Yu Jiang, Yudong Cai, Fengting Bai, Min Qiu, Chen Liang, and Yanshuai Feng are inventors on two patent applications related to this work submitted on 27 December 2024 by Northwest A&F University (Patent Application Nos. 202411951967.8 and 202411951964.4). The other authors declare that they have no competing interests.
Figures




References
-
- Ajmone-Marsan P, Garcia JF, Lenstra JA. On the origin of cattle: how aurochs became cattle and colonized the world. Evol Anthropol 2010;19:148–57.
-
- Bouwman AC, Daetwyler HD, Chamberlain AJ, Ponce CH, Sargolzaei M, Schenkel FS, et al. Meta-analysis of genome-wide association studies for cattle stature identifies common genes that regulate body size in mammals. Nat Genet 2018;50:362–7. - PubMed
-
- Hongo H, Pearson J, Öksüz B, Ilgezdi G. The process of ungulate domestication at Çayönü, Southeastern Turkey: a multidisciplinary approach focusing on Bos sp. and Cervus elaphus. Anthropozoologica 2009;44:63–78.
-
- Vigne JD, Peters J, Helmer D. First Steps of Animal Domestication. Oxford: Oxbow Books; 2005.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

