MicroRNA-Mediated Post-Transcriptional Regulation of Enzymes Involved in Herbicide Resistance in Echinochloa oryzicola (Vasinger) Vasinger
- PMID: 40094587
- PMCID: PMC11901636
- DOI: 10.3390/plants14050719
MicroRNA-Mediated Post-Transcriptional Regulation of Enzymes Involved in Herbicide Resistance in Echinochloa oryzicola (Vasinger) Vasinger
Abstract
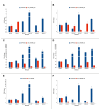
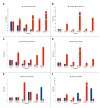
Herbicide resistance is an emerging phytosanitary threat, causing serious yield and economic losses. Although this phenomenon has been widely studied, only recently has the role of epigenetic factors in its occurrence been considered. In the present study, we analyzed the microRNA-mediated regulation in Echinochloa oryzicola (Vasinger) Vasinger (late-watergrass) of the expression of cytochromes P450, glutathione S-transferase (GST), and eIF4B, all of which are enzymes involved in profoxydim (AURA®) detoxification. Before and after profoxydim application, the expression profiles of microRNAs (miRNAs) were selected for their ability to target the genes considered, and their targets were assessed by means of RT-qPCR. Susceptible and resistant biotypes showed different responses to this herbicide. After profoxydim application, in resistant biotypes, osa-miR2099-5p, ath-miR396b, osa-miR395f, osa-miR396a-5p, osa-miR166a-5p, osa-miR166d-5p, gra-miR8759, and gma-miR396f were not triggered, allowing the expression of CYP81A, GSTF1, and eIF4B genes and the herbicide's detoxification. Meanwhile, the transcription of ata-miR166c-5p, ath-miR847, osa-miR5538, and gra-miR7487c was triggered, down-regulating CYP71AK2, CYP72A254, CYP72A122, and EcGST expression. In susceptible biotypes, the herbicide stimulated ata-miR166c-5p, ath-miR847, osa-miR5538, gra-miR7487c, osa-miR166a-5p, and gra-miR8759, down-regulating their respective target genes (CYP72A122, CYP71AK2, EcGST, CYP72A254, CYP81A12, and eIF4B). A better understanding of the role of miRNA-mediated epigenetic regulation in herbicide resistance will be useful in planning more targeted and sustainable methods for controlling this phytosanitary threat.
Keywords: CYP450; CYP81A; GST; eIF4B; late-watergrass; microRNA; profoxydim resistance.
Conflict of interest statement
Authors Marta Guarise, Enrico Gozio, Pietro Zarpellon were employed by the company Agricola 2000 S.c.p.A. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Ente Nazionale Risi. [(accessed on 4 May 2023)]. Available online: www.enterisi.it.
-
- Istat—Istituto Nazionale di Statistica. [(accessed on 6 May 2023)]. Available online: http://dati.istat.it/
-
- GIRE® Italian Herbicide Resistance Working Group, 2023. Database of Herbicide Resistance in Italy. [(accessed on 14 March 2023)]. Available online: http://gire.mlib.cnr.it/
-
- Mascanzoni E., Perego A., Marchi N., Scarabel L., Panozzo S., Ferrero A., Acutis M., Sattin M. Epidemiology and agronomic predictors of herbicide resistance in rice at a large scale. Agron. Sustain. Dev. 2018;38:68. doi: 10.1007/s13593-018-0548-9. - DOI
-
- Herbicide Resistance Action Committee (HRAC) [(accessed on 14 March 2023)]. Available online: https://hracglobal.com/
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

