Integrative Analysis Provides Insights into Genes Encoding LEA_5 Domain-Containing Proteins in Tigernut (Cyperus esculentus L.)
- PMID: 40094764
- PMCID: PMC11902115
- DOI: 10.3390/plants14050762
Integrative Analysis Provides Insights into Genes Encoding LEA_5 Domain-Containing Proteins in Tigernut (Cyperus esculentus L.)
Abstract
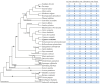
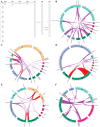
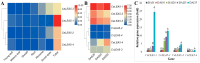
LEA_5 domain-containing proteins constitute a small family of late embryogenesis-abundant proteins that are essential for seed desiccation tolerance and dormancy. However, their roles in non-seed storage organs such as underground tubers are largely unknown. This study presents the first genome-scale analysis of the LEA_5 family in tigernut (Cyperus esculentus L.), a Cyperaceae plant producing desiccation-tolerant tubers. Four LEA_5 genes identified from the tigernut genome are twice of two present in model plants Arabidopsis thaliana and Oryza sativa. A comparison of 86 members from 34 representative plant species revealed the monogenic origin and lineage-specific family evolution in Poales, which includes the Cyperaceae family. CeLEA5 genes belong to four out of five orthogroups identified in this study, i.e., LEA5a, LEA5b, LEA5c, and LEA5d. Whereas LEA5e is specific to eudicots, LEA5b and LEA5d appear to be Poales-specific and LEA5c is confined to families Cyperaceae and Juncaceae. Though no syntenic relationship was observed between CeLEA5 genes, comparative genomics analyses indicated that LEA5b and LEA5c are more likely to arise from LEA5a via whole-genome duplication. Additionally, local duplication, especially tandem duplication, also played a role in the family expansion in Juncus effuses, Joinvillea ascendens, and most Poaceae plants examined in this study. Structural variation (e.g., fragment insertion) and expression divergence of LEA_5 genes were also observed. Whereas LEA_5 genes in A. thaliana, O. sativa, and Zea mays were shown to be preferentially expressed in seeds/embryos, CeLEA5 genes have evolved to be predominantly expressed in tubers, exhibiting seed desiccation-like accumulation during tuber maturation. Moreover, CeLEA5 orthologs in C. rotundus showed weak expression in various stages of tuber development, which may explain the difference in tuber desiccation tolerance between these two close species. These findings highlight the lineage-specific evolution of the LEA_5 family, which facilitates further functional analysis and genetic improvement in tigernut and other species.
Keywords: Cyperaceae; desiccation tolerance; late embryogenesis-abundant protein; phylogenomics; underground tuber; vegetative tissue.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources

