A CPC-shelterin-BTR axis regulates mitotic telomere deprotection
- PMID: 40097392
- PMCID: PMC11914695
- DOI: 10.1038/s41467-025-57456-8
A CPC-shelterin-BTR axis regulates mitotic telomere deprotection
Abstract
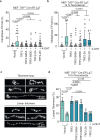
Telomeres prevent ATM activation by sequestering chromosome termini within telomere loops (t-loops). Mitotic arrest promotes telomere linearity and a localized ATM-dependent telomere DNA damage response (DDR) through an unknown mechanism. Using unbiased interactomics, biochemical screening, molecular biology, and super-resolution imaging, we found that mitotic arrest-dependent (MAD) telomere deprotection requires the combined activities of the Chromosome passenger complex (CPC) on shelterin, and the BLM-TOP3A-RMI1/2 (BTR) complex on t-loops. During mitotic arrest, the CPC component Aurora Kinase B (AURKB) phosphorylated both the TRF1 hinge and TRF2 basic domains. Phosphorylation of the TRF1 hinge domain enhances CPC and TRF1 interaction through the CPC Survivin subunit. Meanwhile, phosphorylation of the TRF2 basic domain promotes telomere linearity, activates a telomere DDR dependent on BTR-mediated double Holliday junction dissolution, and leads to mitotic death. We identify that the TRF2 basic domain functions in mitosis-specific telomere protection and reveal a regulatory role for TRF1 in controlling a physiological ATM-dependent telomere DDR. The data demonstrate that MAD telomere deprotection is a sophisticated active mechanism that exposes telomere ends to signal mitotic stress.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures






References
-
- de Lange, T. Shelterin-mediated telomere protection. Annu. Rev. Genet.52, 223–247 (2018). - PubMed
-
- Karlseder, J., Smogorzewska, A. & de Lange, T. Senescence induced by altered telomere state, not telomere loss. Science295, 2446–2449 (2002). - PubMed
-
- d'Adda di Fagagna, F. et al. A DNA damage checkpoint response in telomere-initiated senescence. Nature426, 194–198 (2003). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

