SSUP-72/PINN-1 coordinates RNA-polymerase II 3' pausing and developmental gene expression in C. elegans
- PMID: 40097442
- PMCID: PMC11914089
- DOI: 10.1038/s41467-025-57847-x
SSUP-72/PINN-1 coordinates RNA-polymerase II 3' pausing and developmental gene expression in C. elegans
Abstract
During exit from Caenorhabditis elegans (C. elegans) L1 developmental arrest, a network of growth- and developmental genes is activated, many of which are organized into operons where transcriptional termination is uncoupled from mRNA 3'-end processing. CDK-12-mediated Pol II CTD S2 phosphorylation enhances SL2 trans-splicing at downstream operonic genes, preventing premature termination and ensuring proper gene expression for developmental progression. Using a genetic screen, we identified the SSUP-72/PINN-1 module as a suppressor of defects induced by CDK-12 inhibition. Loss of SSUP-72/PINN-1 bypasses the requirement for CDK-12 in post-embryonic development. Genome-wide analyses reveal that SSUP-72, a CTD S5P phosphatase, affects Pol II 3' pausing and regulates intra-operon termination. Our findings establish SSUP-72/PINN-1 as a key regulator of Pol II dynamics, coordinating operonic gene expression and growth during C. elegans post-embryonic development.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures

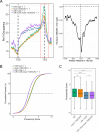
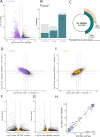
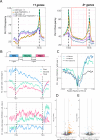
References
-
- Belotserkovskaya, R. & Reinberg, D. Facts about FACT and transcript elongation through chromatin. Curr. Opin. Genet. Dev.14, 139–146 (2004). - PubMed
MeSH terms
Substances
Grants and funding
- FNRS J.0066.16/Fonds De La Recherche Scientifique - FNRS (Belgian National Fund for Scientific Research)
- FNRS T.0112.21/Fonds De La Recherche Scientifique - FNRS (Belgian National Fund for Scientific Research)
- U.N032.22/Fonds De La Recherche Scientifique - FNRS (Belgian National Fund for Scientific Research)
LinkOut - more resources
Full Text Sources
Research Materials

