Light cues drive community-wide transcriptional shifts in the hypersaline South Bay Salt Works
- PMID: 40097557
- PMCID: PMC11914471
- DOI: 10.1038/s42003-025-07855-w
Light cues drive community-wide transcriptional shifts in the hypersaline South Bay Salt Works
Abstract
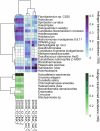
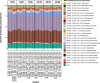
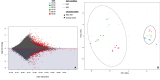
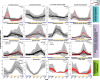
The transition from day to night brings sweeping change to both environments and the organisms within them. Diel shifts in gene expression have been documented across all domains of life but remain understudied in microbial communities, particularly those in extreme environments where small changes may have rippling effects on resource availability. In hypersaline environments, many prominent taxa are photoheterotrophs that rely on organic carbon for growth but can also generate significant ATP via light-powered rhodopsins. Previous research demonstrated a significant response to light intensity shifts in the model halophile Halobacterium salinarum, but these cycles have rarely been explored in situ. Here, we examined genome-resolved differential expression in a hypersaline saltern (water activity (aw) 0.83, total dissolved solids = 250.7 g L-1) throughout a 24-h period. We found increased transcription of genes related to phototrophy and anabolic metabolic processes during the day, while genes related to aerobic respiration and oxidative stress were upregulated at night. Substantiating these results with a chemostat culture of the environmentally abundant halophilic bacterium Salinibacter ruber revealed similar transcriptional upregulation of genes associated with aerobic respiration under dark conditions. These results describe the potential for light-driven changes in oxygen use across both a natural hypersaline environment and a pure culture.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
-
- Mohr, W., Intermaggio, M. P. & LaRoche, J. Diel rhythm of nitrogen and carbon metabolism in the unicellular, diazotrophic cyanobacterium Crocosphaera watsonii WH8501. Environ. Microbiol.12, 412–421 (2010). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

