Comparative phenotypic, physiological, and transcriptomic responses to drought and recovery in two Fraxinus species
- PMID: 40098103
- PMCID: PMC11916329
- DOI: 10.1186/s12870-025-06372-6
Comparative phenotypic, physiological, and transcriptomic responses to drought and recovery in two Fraxinus species
Abstract
Background: This study focused on the drought tolerance and resilience of two ash species: Fraxinus chiisanensis and F. rhynchophylla. These two species are distributed in different habitats, suggesting that they have different levels of drought tolerance. Understanding their response to drought stress, particularly during the seedling stage, is crucial for selecting and developing drought-resistant varieties. This study aimed to compare the phenotypic, physiological, and transcriptomic characteristics of drought-stressed and recovered rewatered plants in a time-course experiment.
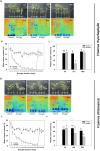
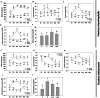
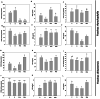
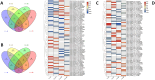
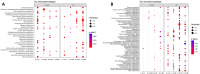
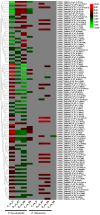
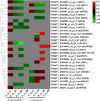
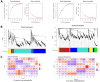
Results: In F. rhynchophylla, drought stress resulted in more severe growth retardation, temperature increase, and a faster decline in the fluorescence response, accompanied by a significant rise in stress indices. However, these reactions recovered quickly after rehydration. In contrast, F. chiisanensis exhibited less growth retardation, a slower decline in fluorescence, and milder increases in stress indices, although many individuals did not fully recover after rehydration. The activity of antioxidant enzymes (SOD, CAT, APX) was more responsive and recovered more efficiently in F. rhynchophylla, while F. chiisanensis had a weaker and delayed response. Transcriptome analysis revealed that photosynthesis and enzyme activity were the most responsive to drought and recovery, as shown by Gene Ontology term analysis. Kyoto Encyclopedia of Genes and Genomes pathway analysis identified common pathways involved in starch and sucrose metabolism and phenylpropanoid biosynthesis in both species. F. rhynchophylla had more differentially expressed genes (DEGs) than F. chiisanensis, particularly on the drought and recovery day 6. Most drought-induced DEGs were restored after rehydration. Commonly associated genes included BGLU and TPS in sugar metabolism; CAT, GSTF, TT7, and HCT in antioxidant enzymes; PYL4 and RR17 in hormone signaling; and ADC1 and ASP3 in proline synthesis.
Conclusions: This study highlights the species-specific characteristics of drought and recovery responses of two Fraxinus species and provides targets for assessing and improving drought tolerance. Moreover, the results of this study provide insights into the physiological and genetic responses of Fraxinus and may guide future research on ash tree stress tolerance.
Keywords: Drought stress5; Fraxinus chiisanensis1; Fraxinus rhynchophylla2; Physiological response4; transcriptome3.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: All the methods involving plants, and their materials complied with relevant institutional, local and national regulations. All plant materials are permitted. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures





References
-
- Bonan GB. Forests and climate change: forcings, feedbacks, and the climate benefits of forests. Science. 2008;320(5882):1444–9. - PubMed
-
- Friedlingstein P, Cox P, Betts R, Bopp L, von Bloh W, Brovkin V, et al. Climate–carbon cycle feedback analysis: results from the C4MIP model intercomparison. J Clim. 2006;19(14):3337–53.
-
- Kennedy D, Swenson S, Oleson KW, Lawrence DM, Fisher R, Lola da Costa AC, et al. Implementing plant hydraulics in the community land model, version 5. J Adv Model Earth Syst. 2019;11(2):485–513.
-
- Trenberth KE, Dai A, Rasmussen RM, Parsons DB. The changing character of precipitation. Bull Amer Meteorol Soc. 2003;84(9):1205–18.
-
- Dai A, Zhao T, Chen J. Climate change and drought. A precipitation and evaporation perspective. Curr Clim Change Rep. 2018;4:301–12.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

