Unravelling the transcriptomic characteristics of bronchoalveolar lavage in post-covid pulmonary fibrosis
- PMID: 40098116
- PMCID: PMC11917041
- DOI: 10.1186/s12920-025-02110-x
Unravelling the transcriptomic characteristics of bronchoalveolar lavage in post-covid pulmonary fibrosis
Abstract
Background: Post-Covid Pulmonary Fibrosis (PCPF) has emerged as a significant global issue associated with a poor quality of life and significant morbidity. Currently, our understanding of the molecular pathways of PCPF is limited. Hence, in this study, we performed whole transcriptome sequencing of the RNA isolated from the bronchoalveolar lavage (BAL) samples of PCPF and compared it with idiopathic pulmonary fibrosis (IPF) and non-ILD (Interstitial Lung Disease) control to understand the gene expression profile and associated pathways.
Methods: BAL samples from PCPF (n = 3), IPF (n = 3), and non-ILD Control (n = 3) (individuals with apparent healthy lung without interstitial lung disease) groups were obtained and RNA were isolated for whole transcriptomic sequencing. Differentially Expressed Genes (DEGs) were determined followed by functional enrichment analysis and qPCR validation.
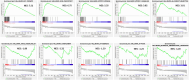
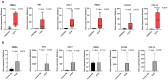
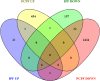
Results: A panel of differentially expressed genes were identified in bronchoalveolar lavage fluid cells (BALF) of PCPF as compare to control and IPF. Our analysis revealed dysregulated pathways associated with cell cycle regulation, immune responses, and neuroinflammatory processes. Real-time validation further supported these findings. The PPI network and module analysis shed light on potential biomarkers and underscore the complex interplay of molecular mechanisms in PCPF. The comparison of PCPF and IPF identified a significant downregulation of pathways that were more prominent in IPF.
Conclusion: This investigation provides crucial insights into the molecular mechanism of PCPF and also outlines avenues for prospective research and the development of therapeutic approaches.
Keywords: BAL; CIBERSORTX; GSEA; IPF; PCPF; RNA-Seq.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This study was performed in accordance with the Declaration of Helsinki. This human study was approved by the Institute Ethics Committee, AIIMS, New Delhi - approval: IEC-388/02.07.2021. All adult participants provided written informed consent to participate in this study. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures




References
-
- Hussain A, Yadav S, Hadda V, Suri TM, Tiwari P, Mittal S et al. Covid-19: a comprehensive review of a formidable foe and the road ahead. Expert Rev Respir Med [Internet]. 2020;14(9):869–79. Available from: https://www.tandfonline.com/doi/full/10.1080/17476348.2020.1782198 - PubMed
-
- Carfì A, Bernabei R, Landi F. Persistent symptoms in patients after acute COVID-19. JAMA [Internet]. 2020;324(6):603. Available from: https://jamanetwork.com/journals/jama/fullarticle/2768351 - PMC - PubMed
-
- Ahmed OF, Kakamad FH, Hama Amin BJ, Abdullah BA, Hassan MN, Salih RQ et al. Post COVID-19 pulmonary complications; a single center experience. Ann Med Surg [Internet]. 2021;72:103052. Available from: https://linkinghub.elsevier.com/retrieve/pii/S2049080121010025 - PMC - PubMed
-
- Polak SB, Van Gool IC, Cohen D, von der Thüsen JH, van Paassen J. A systematic review of pathological findings in COVID-19: a pathophysiological timeline and possible mechanisms of disease progression. Mod Pathol [Internet]. 2020;33(11):2128–38. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0893395222004367 - PMC - PubMed
-
- Udwadia ZF, Koul PA, Richeldi L. Post-COVID lung fibrosis. Lung India [Internet]. 2021;38(Suppl 1):S41–7. Available from: https://journals.lww.com/10.4103/lungindia.lungindia_818_20 - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

