Genome-wide functional annotation of variants: a systematic review of state-of-the-art tools, techniques and resources
- PMID: 40098614
- PMCID: PMC11911558
- DOI: 10.3389/fphar.2025.1474026
Genome-wide functional annotation of variants: a systematic review of state-of-the-art tools, techniques and resources
Abstract
The recent advancement of sequencing technologies marks a significant shift in the character and complexity of the digital genomic data universe, encompassing diverse types of molecular data, screened through manifold technological platforms. As a result, a plethora of fully assembled genomes are generated that span vertically the evolutionary scale. Notwithstanding the tsunami of thriving innovations that accomplish unprecedented, nucleotide-level, structural and functional annotation, an exhaustive, systemic, massive genome-wide functional annotation remains elusive, particularly when the criterion is automation and efficiency in data-agnostic interpretation. The latter is of paramount importance for the elaboration of strategies for sophisticated, data-driven genome-wide annotation, which aim to impart a sustainable and comprehensive systemic approach to addressing whole genome variation. Therefore, it is essential to develop methods and tools that promote systematic functional genomic annotation, with emphasis on mechanistic information exceeding the limits of coding regions, and exploiting the chunks of pertinent information residing in non-coding regions, including promoter and enhancer sequences, non-coding RNAs, DNA methylation sites, transcription factor binding sites, transposable elements and more. This review provides an overview of the current state-of-the-art in genome-wide functional annotation of genetic variation, including existing bioinformatic tools, resources, databases and platforms currently available or reported in the literature. Particular emphasis is placed on the functional annotation of variants that lie outside protein-coding genomic regions (intronic or intergenic), their potential co-localization with regulatory element areas, such as putative non-coding RNA regions, and the assessment of their functional impact on the investigated phenotype. In addition, state-of-the-art tools that leverage data obtained from WGS and GWAS-based analyses are discussed, along with future bioinformatics directions and developments. These future directions emphasize efficient, comprehensive, and largely automated functional annotation of both coding and non-coding genomic variants, as well as their optimal evaluation.
Keywords: GWAS; genome-wide; genomic variation; genotyping; intergenic; variant annotation; whole-exome sequencing; whole-genome sequencing.
Copyright © 2025 Pilalis, Zisis, Andrinopoulou, Karamanidou, Antonara, Stavropoulos and Chatziioannou.
Conflict of interest statement
TK, MA and TS are full-time employees of Pfizer. EP and AC are co-founders and DZ and CA are full-time employees of e-NIOS Applications PC. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision. This research was conducted as a collaboration between e-NIOS Applications and Pfizer. Pfizer is the research sponsor. Pfizer has been involved in the Validation, Writing-review & editing and Project administration.
Figures
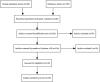
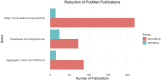
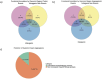
References
Publication types
LinkOut - more resources
Full Text Sources
Research Materials

