Molecular characterization of homogentisate phytyltransferase and methylphytylbenzoquinol methyltransferase genes from olive fruit with regard to the tocopherol content and the response to abiotic stresses
- PMID: 40098644
- PMCID: PMC11911349
- DOI: 10.3389/fpls.2025.1526815
Molecular characterization of homogentisate phytyltransferase and methylphytylbenzoquinol methyltransferase genes from olive fruit with regard to the tocopherol content and the response to abiotic stresses
Abstract
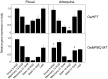
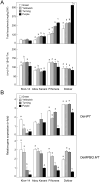
Two cDNA sequences, named OepHPT and OepMPBQ MT, encoding homogentisate phytyltransferase (HPT) and methylphytylbenzoquinol methyltransferase (MPBQ MT), respectively, have been cloned from olive (Olea europaea cv. Picual). Sequence analysis displayed the distinguishing characteristics typical of the HPT and MPBQ MT families and along with phylogenetic analysis indicated that they code for homogentisate phytyltransferase and methylphytylbenzoquinol methyltransferase enzymes, respectively. Transcriptional analysis in distinct olive tissues indicated that expression levels of HPT and MPBQ MT genes are spatially and temporally regulated in a cultivar-dependent manner and together with tocopherol analysis pointed out that both genes participate in the biosynthesis of the tocopherols present in olive mesocarp. These data also suggest that in olive mesocarp, HPT but not MPBQ MT could be implicated in the transcriptional regulation of the tocopherol biosynthetic pathway. In addition, HPT and MPBQ MT transcript levels are regulated by water status, temperature, light, and wounding in the olive fruit mesocarp, suggesting that both genes could be implicated in the abiotic stress response. Overall, this research constitutes a significant advance to elucidate the factors that regulate the tocopherol biosynthesis in olive fruit to obtain virgin olive oils with enhanced α-tocopherol content.
Keywords: HPT; MPBQ MT; Olea europaea; abiotic stresses; gene expression; olive; vitamin E.
Copyright © 2025 Narváez, Hernández, Sicardo, Velázquez-Palmero, Moreda and Martínez-Rivas.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision
Figures






References
-
- Abdallah I. B., Tlili N., Martinez-Force E., Rubio A. G. P., Perez-Camino M. C., Albouchi A., et al. . (2015). Content of carotenoids, tocopherols, sterols, triterpenic and aliphatic alcohols, and volatile compounds in six walnuts (Juglans regia L.) varieties. Food Chem. 173, 972–978. doi: 10.1016/j.foodchem.2014.10.095 - DOI - PubMed
-
- Aparicio R., Harwood J. L. (2013). Handbook of olive oil: analysis and properties (New York, USA: Springer; ).
-
- Baccari S., Elloumi O., Chaari-Rkhis A., Fenollosa E., Morales M., Drira N., et al. . (2020). Linking leaf water potential, photosynthesis and chlorophyll loss with mechanisms of photo- and antioxidant protection in juvenile olive trees subjected to severe drought. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.614144 - DOI - PMC - PubMed
-
- Beltrán G., Jiménez A., Del Río C., Sánchez S., Martínez L., Uceda M., et al. . (2010). Variability of vitamin E in virgin olive oil by agronomical and genetic factors. J. Food Comp. Anal. 23, 633–639. doi: 10.1016/j.jfca.2010.03.003 - DOI
LinkOut - more resources
Full Text Sources

