A conserved motif in Henipavirus P/V/W proteins drives the fibrillation of the W protein from Hendra virus
- PMID: 40100133
- PMCID: PMC11917119
- DOI: 10.1002/pro.70085
A conserved motif in Henipavirus P/V/W proteins drives the fibrillation of the W protein from Hendra virus
Abstract
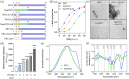
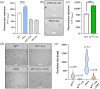
The Hendra (HeV) and Nipah (NiV) viruses are high-priority, biosafety level-4 pathogens that cause fatal neurological and respiratory disease. Their P gene encodes not only the P protein, an essential polymerase cofactor, but also the virulence factors V and W. We previously showed that the W protein of HeV (WHeV) forms amyloid-like fibrils and that one of its subdomains, PNT3, fibrillates in isolation. However, the fibrillation kinetics is much faster in the case of the full-length WHeV compared to PNT3, suggesting that another WHeV region contributes to the fibrillation process. In this work, we identified the region spanning residues 2-110 (PNT1) as the crucial region implicated in WHeV fibrillation. Through site-directed mutagenesis, combined with thioflavin T binding experiments and negative-staining transmission electron microscopy, we showed that a predicted cryptic amyloidogenic region (CAR) within PNT1 is the main driver of fibrillation and deciphered the underlying molecular mechanism. Using FTIR, we showed that PNT1 fibrils are enriched in cross β-sheets. Sequence alignment revealed conservation of the CAR across the Henipavirus genus and enabled the identification of a hitherto never reported pro-amyloidogenic motif. The ability to form fibrils was experimentally shown to be a common property shared by Henipavirus PNT1 proteins. Overall, this study sheds light on the molecular mechanisms underlying WHeV fibrillation and calls for future studies aimed at exploring the relevance of the newly identified pro-amyloidogenic motif as a valuable target for antiviral approaches.
Keywords: FTIR spectroscopy; amyloidogenic regions and motifs; circular dichroism; fibrillation mechanisms; negative‐staining transmission electron microscopy; thioflavin T binding.
© 2025 The Author(s). Protein Science published by Wiley Periodicals LLC on behalf of The Protein Society.
Figures


References
-
- Alberti S, Hyman AA. Biomolecular condensates at the nexus of cellular stress, protein aggregation disease and ageing. Nat Rev Mol Cell Biol. 2021;22:196–213. - PubMed
-
- Basler CF. Nipah and Hendra virus interactions with the innate immune system. Curr Top Microbiol Immunol. 2012;359:123–152. - PubMed
-
- Bignon C, Troilo F, Gianni S, Longhi S. Partner‐mediated polymorphism of an intrinsically disordered protein. J Mol Biol. 2018;430:2493–2507. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

