Reactivation of CTLA4-expressing T cells accelerates resolution of lung fibrosis in a humanized mouse model
- PMID: 40100323
- PMCID: PMC12077895
- DOI: 10.1172/JCI181775
Reactivation of CTLA4-expressing T cells accelerates resolution of lung fibrosis in a humanized mouse model
Abstract
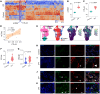
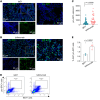
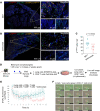
Tissue regenerative responses involve complex interactions between resident structural and immune cells. Recent reports indicate that accumulation of senescent cells during injury repair contributes to pathological tissue fibrosis. Using tissue-based spatial transcriptomics and proteomics, we identified upregulation of the immune checkpoint protein, cytotoxic T lymphocyte-associated protein 4 (CTLA4), on CD8+ T cells adjacent to regions of active fibrogenesis in human idiopathic pulmonary fibrosis and in a repetitive bleomycin lung injury murine model of persistent fibrosis. In humanized CTLA4-knockin mice, treatment with ipilimumab, an FDA-approved drug that targets CTLA4, resulted in accelerated lung epithelial regeneration and diminished fibrosis from repetitive bleomycin injury. Ipilimumab treatment resulted in the expansion of Cd3e+ T cells, diminished accumulation of senescent cells, and robust expansion of type 2 alveolar epithelial cells, facultative progenitor cells of the alveolar epithelium. Ex vivo activation of isolated CTLA4-expressing CD8+ cells from mice with established fibrosis resulted in enhanced cytolysis of senescent cells, suggesting that impaired immune-mediated clearance of these cells contributes to persistence of lung fibrosis in this murine model. Our studies support the concept that endogenous immune surveillance of senescent cells may be essential in promoting tissue regenerative responses that facilitate the resolution of fibrosis.
Keywords: Fibrosis; Immunology; Immunotherapy; Pulmonology; T cells.
Conflict of interest statement
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

