Using Synthetic Health Care Data to Leverage Large Language Models for Named Entity Recognition: Development and Validation Study
- PMID: 40101227
- PMCID: PMC11962312
- DOI: 10.2196/66279
Using Synthetic Health Care Data to Leverage Large Language Models for Named Entity Recognition: Development and Validation Study
Abstract
Background: Named entity recognition (NER) plays a vital role in extracting critical medical entities from health care records, facilitating applications such as clinical decision support and data mining. Developing robust NER models for low-resource languages, such as Estonian, remains a challenge due to the scarcity of annotated data and domain-specific pretrained models. Large language models (LLMs) have proven to be promising in understanding text from any language or domain.
Objective: This study addresses the development of medical NER models for low-resource languages, specifically Estonian. We propose a novel approach by generating synthetic health care data and using LLMs to annotate them. These synthetic data are then used to train a high-performing NER model, which is applied to real-world medical texts, preserving patient data privacy.
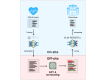
Methods: Our approach to overcoming the shortage of annotated Estonian health care texts involves a three-step pipeline: (1) synthetic health care data are generated using a locally trained GPT-2 model on Estonian medical records, (2) the synthetic data are annotated with LLMs, specifically GPT-3.5-Turbo and GPT-4, and (3) the annotated synthetic data are then used to fine-tune an NER model, which is later tested on real-world medical data. This paper compares the performance of different prompts; assesses the impact of GPT-3.5-Turbo, GPT-4, and a local LLM; and explores the relationship between the amount of annotated synthetic data and model performance.
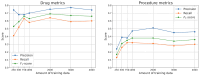
Results: The proposed methodology demonstrates significant potential in extracting named entities from real-world medical texts. Our top-performing setup achieved an F1-score of 0.69 for drug extraction and 0.38 for procedure extraction. These results indicate a strong performance in recognizing certain entity types while highlighting the complexity of extracting procedures.
Conclusions: This paper demonstrates a successful approach to leveraging LLMs for training NER models using synthetic data, effectively preserving patient privacy. By avoiding reliance on human-annotated data, our method shows promise in developing models for low-resource languages, such as Estonian. Future work will focus on refining the synthetic data generation and expanding the method's applicability to other domains and languages.
Keywords: Estonian; LLM; NER; NLP; annotated data; artificial intelligence; clinical decision support; data annotation; data mining; health care data; language model; large language model; machine learning; medical entity; named entity recognition; natural language processing; synthetic data.
©Hendrik Šuvalov, Mihkel Lepson, Veronika Kukk, Maria Malk, Neeme Ilves, Hele-Andra Kuulmets, Raivo Kolde. Originally published in the Journal of Medical Internet Research (https://www.jmir.org), 18.03.2025.
Conflict of interest statement
Conflicts of Interest: None declared.
Figures
Similar articles
-
Improving large language models for clinical named entity recognition via prompt engineering.J Am Med Inform Assoc. 2024 Sep 1;31(9):1812-1820. doi: 10.1093/jamia/ocad259. J Am Med Inform Assoc. 2024. PMID: 38281112 Free PMC article.
-
Prompt Framework for Extracting Scale-Related Knowledge Entities from Chinese Medical Literature: Development and Evaluation Study.J Med Internet Res. 2025 Mar 18;27:e67033. doi: 10.2196/67033. J Med Internet Res. 2025. PMID: 40100267 Free PMC article.
-
Improving entity recognition using ensembles of deep learning and fine-tuned large language models: A case study on adverse event extraction from VAERS and social media.J Biomed Inform. 2025 Mar;163:104789. doi: 10.1016/j.jbi.2025.104789. Epub 2025 Feb 7. J Biomed Inform. 2025. PMID: 39923968
-
Large Language Model Applications for Health Information Extraction in Oncology: Scoping Review.JMIR Cancer. 2025 Mar 28;11:e65984. doi: 10.2196/65984. JMIR Cancer. 2025. PMID: 40153782 Free PMC article.
-
Natural Language Processing for Digital Health in the Era of Large Language Models.Yearb Med Inform. 2024 Aug;33(1):229-240. doi: 10.1055/s-0044-1800750. Epub 2025 Apr 8. Yearb Med Inform. 2024. PMID: 40199310 Free PMC article. Review.
Cited by
-
Large Language Model Synergy for Ensemble Learning in Medical Question Answering: Design and Evaluation Study.J Med Internet Res. 2025 Jul 14;27:e70080. doi: 10.2196/70080. J Med Internet Res. 2025. PMID: 40658884 Free PMC article.
References
-
- Li I, Pan J, Goldwasser J, Verma N, Wong W, Nuzumlalı MY, Rosand B, Li Y, Zhang M, Chang D, Taylor Ra, Krumholz Hm, Radev D. Neural natural language processing for unstructured data in electronic health records: a review. Comput Sci Rev. 2022 Nov;46:100511. doi: 10.1016/j.cosrev.2022.100511. https://www.sciencedirect.com/science/article/pii/S1574013722000454 - DOI
-
- Torge S, Politov A, Lehmann C, Saffar B, Tao Z. Named entity recognition for low-resource languages - profiting from language families. Proceedings of the 9th Workshop on Slavic Natural Language Processing 2023 (SlavicNLP 2023); May 6, 2023; Dubrovnik, Croatia. Association for Computational Linguistics; 2023. pp. 1–10. https://aclanthology.org/2023.bsnlp-1.1 - DOI
-
- Cotterell R, Duh K. Low-resource named entity recognition with cross-lingual, character-level neural conditional random fields. Proceedings of the Eighth International Joint Conference on Natural Language Processing (Volume 2: Short Papers); November 27 - December 1, 2017; Taipei, Taiwan. ACM; 2017. pp. 91–96. https://aclanthology.org/I17-2016 - DOI
-
- Simpson E, Brown R, Sillence E, Coventry L, Lloyd K, Gibbs J, Tariq S, Durrant AC. Understanding the barriers and facilitators to sharing patient-generated health data using digital technology for people living with long-term health conditions: a narrative review. Front Public Health. 2021;9:641424. doi: 10.3389/fpubh.2021.641424. https://europepmc.org/abstract/MED/34888271 - DOI - PMC - PubMed
-
- Szarvas G, Farkas R, Busa-Fekete R. State-of-the-art anonymization of medical records using an iterative machine learning framework. J Am Med Inform Assoc. 2007;14(5):574–580. doi: 10.1197/j.jamia.M2441. https://europepmc.org/abstract/MED/17823086 14/5/574 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources