Large-scale computational modelling of H5 influenza variants against HA1-neutralising antibodies
- PMID: 40101386
- PMCID: PMC11960665
- DOI: 10.1016/j.ebiom.2025.105632
Large-scale computational modelling of H5 influenza variants against HA1-neutralising antibodies
Abstract
Background: The United States Department of Agriculture has recently released reports that show samples collected from 2022 to 2025 of highly pathogenic avian influenza (H5N1) have been detected in mammals and birds. Up to February 2025, the United States Centres for Disease Control and Prevention reports that there have been 67 humans infected with H5N1 since 2024 with 1 death. The broader potential impact on human health remains unclear.
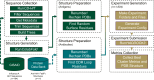
Methods: In this study, we computationally model 1804 protein complexes consisting of various H5 isolates from 1959 to 2024 against 11 haemagglutinin domain 1 (HA1)-neutralising antibodies. This was performed using AI-based protein folding and physics-based simulations of the antibody-antigen interactions. We analysed binding affinity changes over time and across various antibodies using multiple biochemical and biophysical binding metrics.
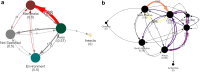
Findings: This study shows a trend of weakening binding affinity of existing antibodies against H5 isolates over time, indicating that the H5N1 virus is evolving immune escape from our therapeutic and immunological defences. We also found that based on the wide variety of host species and geographic locations in which H5N1 was observed to have been transmitted from birds to mammals, there is not a single central reservoir host species or location associated with H5N1's spread.
Interpretation: These results indicate that the virus has potential to move from epidemic to pandemic status. This study illustrates the value of high-performance computing to rapidly model protein-protein interactions and viral genomic sequence data at-scale for functional insights into medical preparedness.
Funding: No external funding was used in this study.
Keywords: Antibodies; Avian influenza; Docking; Protein modelling; Zoonosis.
Copyright © 2025 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests Author CTF is the owner of Tuple, LLC, a biotechnology consulting firm. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. Author DJ is the director of the UNC Charlotte Center for Computational Intelligence to Predict Health and Environmental Risks (CIPHER), which is funded by an Ignite grant from the UNC Charlotte Division of Research.
Figures




References
-
- USDA Animal and Plant Health Inspection Service . 2024. USDA APHIS livestock and poultry health: avian influenza high pathogenicity avian influenza detections in mammals.https://www.aphis.usda.gov/livestock-poultry-disease/avian/avian-influen...
-
- CDC . 2024. H5 bird flu: current situation.https://www.cdc.gov/bird-flu/situation-summary/index.html
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

