Limitations of sequence dissimilarity as a predictor of prokaryotic lineage
- PMID: 40101780
- PMCID: PMC11919493
- DOI: 10.1098/rsob.240302
Limitations of sequence dissimilarity as a predictor of prokaryotic lineage
Abstract
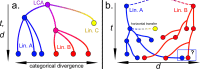
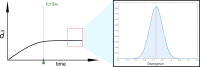
The molecular clock rests upon the assumption that the observed changes among sequences capture the differentiation of lineages, or kinship, as dissimilarity increases with time. Although it has been questioned over the years, this paradigmatic principle continues to underlie the idea that the polymorphic space of a gene is so vast that it is unattainable in evolutionary time. Thus, the molecular clock has been used to obtain taxonomic annotations, proving to be very effective at delivering testable results. In this article, however, we ask how often this assumption leads to inaccuracies when inferring the lineage of prokaryotic genes. Thus, we open an interesting discussion by simulating, in realistic scenarios, the critical times in which specific 5S rRNA sequences of two distant lineages are exhausting the polymorphic space. We contend that certain genes in one lineage will become increasingly similar to those in another over time, as the space for new variants is finite, mimicking phylogenetic features by convergence or by chance, without implying true kinship.
Keywords: gene polymorphism; molecular clock; molecular evolution; phylogenetic time; prokaryotic evolution; sequence dissimilarity.
Conflict of interest statement
We declare we have no competing interests.
Figures


References
-
- Sewall W. 1941. The material basis of evolution. Sci. Mon. 53, 165–170.
-
- Zuckerkandl E, Pauling L. 1965. Evolutionary divergence and convergence in proteins. Piscataway, NJ: Elsevier. ( 10.1016/B978-1-4832-2734-4.50017-6) - DOI
-
- Kimura M. 1983. The neutral theory of molecular evolution. Cambridge, UK: Cambridge University Press.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
