Comprehensive sequencing profile and functional analysis of IsomiRs in human pancreatic islets and beta cells
- PMID: 40102237
- PMCID: PMC12069488
- DOI: 10.1007/s00125-025-06397-4
Comprehensive sequencing profile and functional analysis of IsomiRs in human pancreatic islets and beta cells
Abstract
Aims/hypothesis: MiRNAs regulate gene expression, influencing beta cell function and pathways. Isoforms of miRNA (isomiRs), sequence variants of miRNAs with post-transcriptional modifications, exhibit cell-type-specific expression and functions. Despite their biological significance, a comprehensive isomiR profile in human pancreatic islets and beta cells remains unexplored. This study aims to profile isomiR expression in four beta cell sources: (1) laser capture microdissected human islets (LCM-HI); (2) collagenase-isolated human islets (CI-HI); (3) sorted beta cells; and (4) the EndoC-βH1 beta cell line, and to investigate their potential role in beta cell function.
Methods: Small RNA-seq and/or small RNA dataset analysis was conducted on human pancreatic islets and beta cells. Data were processed using the sRNAbench bioinformatics pipeline to classify isomiRs based on sequence variations. A beta cell-specific isomiR signature was identified via cross-validation across datasets. Correlations between LCM-HI isomiR expression and in vivo clinical parameters were analysed using regression models. Functional validation of isomiR-411-5p-Ext5p(+1) was performed via overexpression in EndoC-βH1 cells and CI-HI, followed by glucose-stimulated insulin secretion (GSIS) assays and/or transcriptomic analysis.
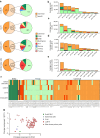
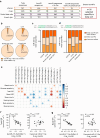
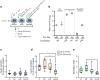
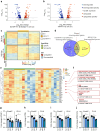
Results: IsomiRs constituted 59.2 ± 1.9% (LCM-HI), 59.6 ± 2.4% (CI-HI), 42.3 ± 7.2% (sorted beta cells) and 43.8 ± 1.2% (EndoC-βH1) of total miRNA reads (data represented as mean ± SD), with 3' end trimming (Trim3p) being the predominant modification. A beta cell-specific isomiR signature of 30 sequences was identified, with isomiR-411-5p-Ext5p(+1) showing a significant inverse correlation with basal insulin secretion (p=0.0009, partial R2=0.68) and total insulin secretion (p=0.005, partial R2=0.54). Overexpression of isomiR-411-5p-Ext5p(+1), but not of its canonical counterpart, importantly reduced GSIS by 51% ( ± 15.2%; mean ± SD) (p=0.01) in EndoC-βH1 cells. Transcriptomic analysis performed in EndoC-βH1 cells and CI-HI identified 47 genes significantly downregulated by isomiR-411-5p-Ext5p(+1) (false discovery rate [FDR]<0.05) but not by the canonical miRNA, with enriched pathways related to Golgi vesicle biogenesis (FDR=0.017) and trans-Golgi vesicle budding (FDR=0.018). TargetScan analysis confirmed seed sequence-dependent target specificity for 81 genes uniquely regulated by the isomiR (p=1.1 × 10⁻⁹).
Conclusions/interpretation: This study provides the first comprehensive isomiR profiling in human islets and beta cells, revealing their substantial contribution to miRNA regulation. IsomiR-411-5p-Ext5p(+1) emerges as a distinct key modulator of insulin secretion and granule dynamics in beta cells. These findings highlight isomiRs as potential biomarkers and therapeutic targets for diabetes, warranting further exploration of their roles in beta cell biology.
Keywords: Beta cell; Beta cell function; Human pancreatic islets; Insulin secretion; IsomiRs; MicroRNAs; Non-coding RNAs.
© 2025. The Author(s).
Conflict of interest statement
Acknowledgements: The secretarial help of A. Mechini and M. Prencipe was highly appreciated. BioRender.com images tool was used to generate the Graphical abstract. Data availability: Raw and analysed data are available from the corresponding author upon request. Funding: This work is supported by the European Union (EU) within the Italian Ministry of University and Research (MUR) PNRR ‘National Center for Gene Therapy and Drugs based on RNA Technology’ (Project No. CN00000041 CN3 Spoke #5 ‘Inflammatory and Infectious Diseases’), and by the Innovative Medicines Initiative 2 (IMI2) Joint Undertaking under grant agreement No.115797 INNODIA and No.945268 INNODIA HARVEST. This joint undertaking receives support from the Union’s Horizon 2020 research and innovation programme and EFPIA, Breakthrough T1D (former JDRF) and The Leona M. and Harry B. Helmsley Charitable Trust). The work is also supported by the Italian Ministry of Health through ‘Bando Ricerca Finalizzata 2018’ GR-2018-12365577: ‘The study of human pancreatic islet cell plasticity to predict diabetes onset, progression and personalize therapy’. GS, AP and TM are supported by the Italian Ministry of University and Research (PNRR-PRIN2022 No. P2022EB5B8 and PRIN2022 No.2022FRBXHY). GS is supported by the University of Siena within F-CUR funding program Grant No. 2268-2022-SG-PSR2021-FCUR_001. FD was supported by the Italian Ministry of University and Research with the project PNC 0000001 D3 4 Health, the National Plan for Complementary Investments to the NRRP, funded by the NextGenerationEU, and by the Italian Ministry of Health 'Multidisciplinary and Interregional Hub for Research and Clinical Experimentation To Combat Pandemics and Antibiotic Resistance' (project T4-AN-07, PAN-HUB). Authors’ relationships and activities: The authors declare that there are no relationships or activities that might bias, or be perceived to bias, their work. Contribution statement: SA, EA and GEG contributed to the conceptualisation, supervision and coordination of the study, the design of the methodology, conducted the investigation and implemented the bioinformatics and statistical analysis, composed the figures and wrote and reviewed the manuscript. DF, GL, AMo, AFB, EP, LN and CF conducted the experiments and the analyses and interpreted the data. MB contributed to the design of the methodology. CG contributed to the investigation and to the design of the methodology. GQ and VT contributed to the investigation and to the design of the methodology. GDG, LS and GC contributed to the investigation and to the design of the methodology. AMa contributed to the investigation. AG and TM contributed to the investigation, to the design of the methodology and helped acquire funding for the research. AP contributed to the investigation to the design of the methodology and helped acquire funding for the research. RR contributed to the investigation and to the design of the methodology. FD and GS contributed to the conceptualisation, supervision and coordination of the study and the design of the methodology, wrote and reviewed the manuscript and helped acquire funding for the research. FD and GS are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of their analysis. All authors substantially contributed to the conception or design of the work, or acquisition, analysis or interpretation of data of the work. All authors critically reviewed the manuscript and approved the final version.
Figures
References
-
- Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75:843–854. 10.1016/0092-8674(93)90529-y - PubMed
MeSH terms
Substances
Grants and funding
- Project No. CN00000041/Italian Ministry of University and Research (MUR) PNRR "National Center for Gene Therapy and Drugs based on RNA Technology" , CN3 Spoke #5 "Inflammatory and Infectious Diseases"
- 2017KAM2R5_003/Italian Ministry of University and Research
- GR-2018-12365577/Bando Ricerca Finalizzata 2018, Italian Ministry of Health
- 115797 INNODIA/Innovative Medicines Initiative 2 (IMI2) Joint Undertaking
- 945268 INNODIA HARVEST/Innovative Medicines Initiative 2 (IMI2) Joint Undertaking
LinkOut - more resources
Full Text Sources

