In vitro competition with Bifidobacterium strains impairs potentially pathogenic growth of Clostridium perfringens on 2'-fucosyllactose
- PMID: 40102238
- PMCID: PMC11956901
- DOI: 10.1080/19490976.2025.2478306
In vitro competition with Bifidobacterium strains impairs potentially pathogenic growth of Clostridium perfringens on 2'-fucosyllactose
Abstract
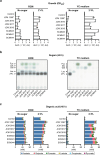
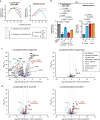
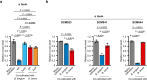
Fortifying infant formula with human milk oligosaccharides, such as 2'-fucosyllactose (2'-FL), is a global trend. Previous studies have shown the inability of pathogenic gut microbes to utilize 2'-FL. However, the present study demonstrates that the type strain (JCM 1290T) of Clostridium perfringens, a pathobiont species often more prevalent and abundant in the feces of C-section-delivered infants, exhibits potentially pathogenic growth on 2'-FL. The expression of genes for α-toxin, an activator of NLRP3 inflammasome, and ethanolamine ammonia-lyase, a factor responsible for the progression of gas gangrene, was significantly upregulated during 2'-FL assimilation compared to growth on lactose. However, colony-forming unit of C. perfringens JCM 1290T markedly decreased when co-cultivated with selected strains of Bifidobacterium, a taxon frequently detected in the breastfed infant gut. Moreover, during co-cultivation, the expression of virulence-related genes, including the gene for perfringolysin O - another activator of NLRP3 inflammasome - were significantly downregulated, while the lactate oxidation genes were upregulated. This can occur through two different mechanisms: direct competition for 2'-FL between the two organisms, or cross-feeding of lactose, released from 2'-FL by C. perfringens JCM 1290T, to Bifidobacterium. Attenuation of α-toxin production by the selected Bifidobacterium strains was observed to varying extents in 2'-FL-utilizing C. perfringens strains clinically isolated from healthy infants. Our results warrant detailed in vivo studies using animal models with dysbiotic microbiota dominated by various types of C. perfringens strains to further validate the safety of 2'-FL for clinical interventions, particularly on vulnerable preterm infants.
Keywords: 2'-fucosyllactose; Bifidobacteria; Clostridium perfringens; α-toxin.
Conflict of interest statement
Employment of MS (since October, 2020), MNO (since April, 2021), and TomK (from April, 2021 to March, 2022) at Kyoto University is in part supported by Morinaga Milk Industry Co., Ltd. RM, KY, J-ZX, and TO are employees of Morinaga Milk Industry Co., Ltd. DAR is a co-founder of PhenoBiome Inc.
Figures


References
-
- McGuire M, Meehan CL, McGuire MA, Williams JE, Foster J, Sellen DW, Kamau-Mbuthia EW, Kamundia EW, Mbugua S, Moore SE, et al. What’s normal? Oligosaccharide concentrations and profiles in milk produced by healthy women vary geographically. Am J Clin Nutr. 2017;105(5):1086–19. doi: 10.3945/ajcn.116.139980. - DOI - PMC - PubMed
-
- V CG, Zampini L, Galeazzi T, Facinelli B, Ferrante L, Capretti R, Orazio G. Human milk oligosaccharides inhibit the adhesion to caco-2 cells of diarrheal pathogens Escherichia coli, Vibrio cholerae, and Salmonella fyris. Pediatr Res. 2006;59(3):377–382. doi: 10.1203/01.pdr.0000200805.45593.17. - DOI - PubMed
-
- Sodhi CP, Wipf P, Yamaguchi Y, Fulton WB, Kovler M, Niño DF, Zhou Q, Banfield E, Werts AD, Ladd MR, et al. The human milk oligosaccharides 2’-fucosyllactose and 6’-sialyllactose protect against the development of necrotizing enterocolitis by inhibiting toll-like receptor 4 signaling. Pediatr Res. 2021;89(1):91–101. doi: 10.1038/s41390-020-0852-3. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
