Genomic structural variation in an alpha/beta hydrolase triggers hybrid necrosis in wheat
- PMID: 40102399
- PMCID: PMC11920055
- DOI: 10.1038/s41467-025-57750-5
Genomic structural variation in an alpha/beta hydrolase triggers hybrid necrosis in wheat
Abstract
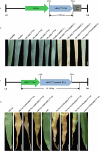
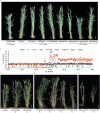
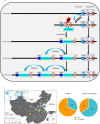
Hybrid necrosis, a century-old mystery in wheat, is caused by complementary genes Ne1 and Ne2. Ne2, encoding a nucleotide-binding leucine-rich repeat (NLR) immune receptor, has been cloned, yet Ne1 remains elusive. Here, we report that Ne1, which encodes an alpha/beta hydrolase (ABH) protein generated by structural variation, triggers hybrid necrosis with Ne2 by activating autoimmune responses. We further verify that not only allelic variation but also copy number variation (CNV) of Ne1 are pivotal for hybrid necrosis diversity in wheat. Ne1 likely originates from wild emmer wheat, potentially through duplication and ectopic recombination events. Unlike Ne2, which is frequently selected for rust resistance in wheat breeding, the lower prevalence of Ne1 in modern wheat cultivars is attributed to its association with hybrid necrosis. Altogether, these findings illuminate the co-evolution of the NLR/ABH gene pair in plant development and innate immunity, offering potential benefits for wheat breeding.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



Similar articles
-
Fine mapping and distribution analysis of hybrid necrosis genes Ne1 and Ne2 in wheat in China.Theor Appl Genet. 2022 Apr;135(4):1177-1189. doi: 10.1007/s00122-021-04023-6. Epub 2022 Jan 28. Theor Appl Genet. 2022. PMID: 35088104
-
Ne2, a typical CC-NBS-LRR-type gene, is responsible for hybrid necrosis in wheat.New Phytol. 2021 Oct;232(1):279-289. doi: 10.1111/nph.17575. Epub 2021 Jul 21. New Phytol. 2021. PMID: 34160845
-
Fine mapping of Ne1, the hybrid necrosis gene complementary to Ne2 in common wheat (Triticum aestivum L.).Theor Appl Genet. 2021 Sep;134(9):2813-2821. doi: 10.1007/s00122-021-03860-9. Epub 2021 May 22. Theor Appl Genet. 2021. PMID: 34023915
-
Recent advances of NLR receptors in vegetable disease resistance.Plant Sci. 2024 Nov;348:112224. doi: 10.1016/j.plantsci.2024.112224. Epub 2024 Aug 12. Plant Sci. 2024. PMID: 39142606 Review.
-
NLR immune receptors and diverse types of non-NLR proteins control race-specific resistance in Triticeae.Curr Opin Plant Biol. 2021 Aug;62:102053. doi: 10.1016/j.pbi.2021.102053. Epub 2021 May 28. Curr Opin Plant Biol. 2021. PMID: 34052730 Review.
References
-
- Bomblies, K. & Weigel, D. Hybrid necrosis: autoimmunity as a potential gene-flow barrier in plant species. Nat. Rev. Genet.8, 382–393 (2007). - PubMed
-
- Li, L. & Weigel, D. One hundred years of hybrid necrosis: hybrid autoimmunity as a window into the mechanisms and evolution of plant-pathogen interactions. Annu. Rev. Phytopathol.59, 213–237 (2021). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

