Unraveling the molecular mechanism of polysaccharide lyases for efficient alginate degradation
- PMID: 40102416
- PMCID: PMC11920209
- DOI: 10.1038/s41467-025-56754-5
Unraveling the molecular mechanism of polysaccharide lyases for efficient alginate degradation
Abstract
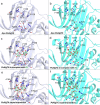
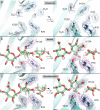
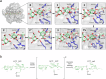
Alginate lyases (ALs) catalyze the depolymerization of brown macroalgae alginates, widely used naturally occurring polysaccharides. Their molecular reaction mechanism remains elusive due to the lack of catalytically competent Michaelis-Menten-like complex structures. Here, we provide structural snapshots and dissect the mechanism of mannuronan-specific ALs from family 7 polysaccharide lyases (PL7), employing time-resolved NMR, X-ray, neutron crystallography, and QM/MM simulations. We reveal the protonation state of critical active site residues, enabling atomic-level analysis of the reaction coordinate. Our approach reveals an endolytic and asynchronous syn β-elimination reaction, with Tyr serving as both Brønsted base and acid, involving a carbanion-type transition state. This study not only reconciles previous structural and kinetic discrepancies, but also establishes a comprehensive PL reaction mechanism which is most likely applicable across all enzymes of the PL7 family as well as other PL families.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures

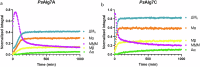



Similar articles
-
Structure of a PL17 family alginate lyase demonstrates functional similarities among exotype depolymerases.J Biol Chem. 2014 Mar 21;289(12):8645-55. doi: 10.1074/jbc.M113.531111. Epub 2014 Jan 29. J Biol Chem. 2014. PMID: 24478312 Free PMC article.
-
Structure/activity relationships of two alginate lyases from Flavobacterium spp. and their potential application in detergents.Int J Biol Macromol. 2025 May;310(Pt 4):143524. doi: 10.1016/j.ijbiomac.2025.143524. Epub 2025 Apr 25. Int J Biol Macromol. 2025. PMID: 40288722
-
Insights into the mechanism of substrate specificity in a novel PL15_3 subfamily oligo-alginate lyase VBAly15A.Appl Environ Microbiol. 2025 Mar 19;91(3):e0235124. doi: 10.1128/aem.02351-24. Epub 2025 Feb 27. Appl Environ Microbiol. 2025. PMID: 40013786 Free PMC article.
-
Diversity of Three-Dimensional Structures and Catalytic Mechanisms of Alginate Lyases.Appl Environ Microbiol. 2018 Jan 17;84(3):e02040-17. doi: 10.1128/AEM.02040-17. Print 2018 Feb 1. Appl Environ Microbiol. 2018. PMID: 29150496 Free PMC article. Review.
-
Alginate lyase: Review of major sources and classification, properties, structure-function analysis and applications.Bioengineered. 2015;6(3):125-31. doi: 10.1080/21655979.2015.1030543. Epub 2015 Apr 1. Bioengineered. 2015. PMID: 25831216 Free PMC article. Review.
Cited by
-
Modeling catalytic reaction on ligand-protected metal nanoclusters.Chem Sci. 2025 May 19;16(26):12080-12086. doi: 10.1039/d5sc00421g. eCollection 2025 Jul 2. Chem Sci. 2025. PMID: 40474957 Free PMC article.
-
Exploring the Catalytic Mechanisms of a Newly Identified Salt-Activated Alginate Lyase from Pseudoalteromonas carrageenovora ASY5.Mar Drugs. 2025 Jun 15;23(6):254. doi: 10.3390/md23060254. Mar Drugs. 2025. PMID: 40559663 Free PMC article.
References
-
- Aarstad, O. A., Tøndervik, A., Sletta, H. & Skjåk-Bræk, G. Alginate sequencing: an analysis of block distribution in alginates using specific alginate degrading enzymes. Biomacromolecules13, 106–116 (2012). - PubMed
-
- Kloareg, B. & Quatrano, R. S. Structure of the cell walls of marine algae and ecophysiological functions of the matrix polysaccharides. Oceanogr. Mar. Biol. Annu. Rev.26, 259–315 (1988).
-
- Nøkling-Eide, K. et al. Acid preservation of cultivated brown algae Saccharina latissima and Alaria esculenta and characterization of extracted alginate and cellulose. Algal Res.71, 103057 (2023).
-
- Lu, S., Na, K., Wei, J., Zhang, L. & Guo, X. Alginate oligosaccharides: the structure-function relationships and the directional preparation for application. Carbohydr. Polym.284, 119225 (2022). - PubMed
MeSH terms
Substances
Grants and funding
- 315385/Norges Forskningsråd (Research Council of Norway)
- 226244/Norges Forskningsråd (Research Council of Norway)
- 294946/Norges Forskningsråd (Research Council of Norway)
- DFF170746/Det Frie Forskningsråd (Danish Council for Independent Research)
- 2021-SGR-00680/Government of Catalonia | Agència de Gestió d'Ajuts Universitaris i de Recerca (Agency for Management of University and Research Grants)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

