Chromosome-scale assembly of the Xenocypris davidi using PacBio HiFi reads and Hi-C technologies
- PMID: 40102422
- PMCID: PMC11920407
- DOI: 10.1038/s41597-025-04800-8
Chromosome-scale assembly of the Xenocypris davidi using PacBio HiFi reads and Hi-C technologies
Abstract
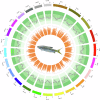
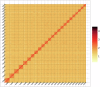
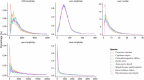
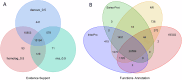
Xenocypris davidi is a benthic fish species widely distributed in the water systems south of the Yellow River in China, playing a significant role in aquatic ecosystems. Despite its ecological and economic importance, genomic resources for X. davidi are limited, hindering a comprehensive understanding of its evolutionary adaptations and genetic improvements. This study presents the first chromosome-level genome assembly of X. davidi, utilizing PacBio long-reads, Illumina short reads, and Hi-C sequencing data. The genome assembly spans 1.05 Gb with a scaffold N50 length of 33.99 Mb, and 95.12% of the genome sequence was successfully anchored onto 24 pseudochromosomes. We identified 27,360 protein-coding genes, of which 26,672 were functionally annotated. This genome sequence provides a valuable resource for exploring the molecular basis of agronomic traits in X. davidi and will facilitate its genetic enhancement.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures
References
-
- Ding, D. Introduction to Four Species of Culter Fish. Hunan Agricuture2, 30+24 (2013).
-
- Xu, D. A Preliminary Analysis on the Food Habie of Xenocypris davidi Bleeker in Reservoir Guangting. Acta Hydrobiologica Sinica01, 43–53 (1988).
-
- Tang, Y. et al. Grazing Effects of Xenocypris davidi Bleeker (Cyprinidae, Cypriniformes) on Filamentous Algae and the Consequent Effects on Intestinal Microbiota. Aquaculture Research2023, 1–14 (2023).
-
- Li, C. et al. Research on the Dietary Protein and Fat Requirements of Parental Xenocypris davidi. Jiangsu Agricultural Sciences47, 220–223 (2019).
-
- Li, C. et al. Effects of Different Levels of Bee Pollen in Feed on Reproductive Performance of Xenocypris davidi Bleeker. Agricultural Science and Technology20, 48–52 (2019).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

