Metabolomics strategy for diagnosing urinary tract infections
- PMID: 40102424
- PMCID: PMC11920235
- DOI: 10.1038/s41467-025-57765-y
Metabolomics strategy for diagnosing urinary tract infections
Abstract
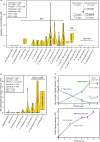
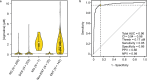
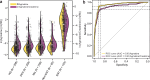
Metabolomics has emerged as a mainstream approach for investigating complex metabolic phenotypes but has yet to be integrated into routine clinical diagnostics. Metabolomics-based diagnosis of urinary tract infections (UTIs) is a logical application of this technology since microbial waste products are concentrated in the bladder and thus could be suitable markers of infection. We conducted an untargeted metabolomics screen of clinical specimens from patients with suspected UTIs and identified two metabolites, agmatine, and N6-methyladenine, that are predictive of culture-positive samples. We developed a 3.2-min LC-MS assay to quantify these metabolites and showed that agmatine and N6-methyladenine correctly identify UTIs caused by 13 Enterobacterales species and 3 non-Enterobacterales species, accounting for over 90% of infections (agmatine AUC > 0.95; N6-methyladenine AUC > 0.89). These markers were robust predictors across two blinded cohorts totaling 1629 patient samples. These findings demonstrate the potential utility of metabolomics in clinical diagnostics for rapidly detecting UTIs.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: Drs. Lewis, Gregson, and Mr. Groves are authors of a patent relating to the use of LC-MS for detecting urinary tract infections. These authors have a relationship with Rapid Infection Diagnostics Inc., a company that is developing commercial microbiology diagnostic tools. No other competing interests were declared.
Figures

References
-
- Fernández-García, M., Rojo, D., Rey-Stolle, F., García, A. & Barbas, C. Metabolomic-based methods in diagnosis and monitoring infection progression. In: Metabolic Interaction in Infection (eds. Silvestre, R. & Torrado, E.) 283–315 (Springer International Publishing, Cham, 2018). 10.1007/978-3-319-74932-7_7. - PMC - PubMed
-
- Satink, H. P. W. M. et al. Microbial influences on urinary polyamine excretion. Clin. Chim. Acta179, 305–314 (1989). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

