The primate-specific Nedd4-1(NE) localizes to late endosomes in response to amino acids to suppress autophagy
- PMID: 40102426
- PMCID: PMC11920435
- DOI: 10.1038/s41467-025-57944-x
The primate-specific Nedd4-1(NE) localizes to late endosomes in response to amino acids to suppress autophagy
Abstract
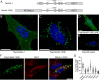
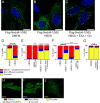
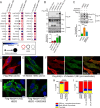
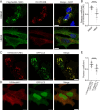
The ubiquitin ligase Nedd4 (Nedd4-1), comprised of C2-WW(n)-HECT domains, regulates protein trafficking. We recently described a primate-specific Nedd4-1 splice isoform with an extended N-terminus replacing the C2 domain, called Nedd4-1(NE). Here, we show that while canonical Nedd4-1 is primarily localized to the cytosol, Nedd4-1(NE) localizes to late endosomes. This localization is mediated by the NE region, is dependent on amino acid availability, is independent of mTORC1, and is inhibited by the autophagy inducer IKKβ. We further demonstrate that VPS16B, which regulates late endosome to lysosome maturation, is a unique Nedd4-1(NE) substrate that co-localizes with Nedd4-1(NE) in the presence of nutrients. Importantly, a potentially pathogenic homozygous variant identified in the NE region (E70Q) of a patient with lymphangiectasia and protein-losing enteropathy leads to reduced VPS16B ubiquitination by Nedd4-1(NE). Finally, we report that Nedd4-1(NE) inhibits autophagy, likely by disrupting late endosome to autophagosome maturation. This work identified an mTORC1-independent, IKK-driven mechanism to regulate Nedd4-1(NE) localization to late endosomes in primates in response to nutrient availability, and uncovered suppression of autophagy by this ubiquitin ligase.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures

References
-
- Hershko, A. & Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem.67, 425–479 (1998). - PubMed
-
- Rotin, D. & Prag, G. Physiological functions of the ubiquitin ligases Nedd4-1 and Nedd4-2. Physiol. (Bethesda)39, 18–29 (2024). - PubMed
-
- Plant, P. J., Yeger, H., Staub, O., Howard, P. & Rotin, D. The C2 domain of the ubiquitin protein ligase Nedd4 mediates Ca2+-dependent plasma membrane localization. J. Biol. Chem.272, 32329–32336 (1997). - PubMed
-
- Wiesner, S. et al. Autoinhibition of the HECT-type ubiquitin ligase Smurf2 through its C2 domain. Cell130, 651–662 (2007). - PubMed
-
- Mari, S. et al. Structural and functional framework for the autoinhibition of Nedd4-family ubiquitin ligases. Structure22, 1639–1649 (2014). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

