Orchestrating epigenetics: a comprehensive review of the methyltransferase SETD6
- PMID: 40102573
- PMCID: PMC11958702
- DOI: 10.1038/s12276-025-01423-2
Orchestrating epigenetics: a comprehensive review of the methyltransferase SETD6
Abstract
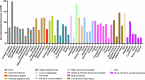
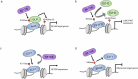
Transcription is regulated by an intricate and extensive network of regulatory factors that impinge upon target genes. This process involves crosstalk between a plethora of factors that include chromatin structure, transcription factors and posttranslational modifications (PTMs). Among PTMs, lysine methylation has emerged as a key transcription regulatory PTM that occurs on histone and non-histone proteins, and several enzymatic regulators of lysine methylation are attractive targets for disease intervention. SET domain-containing protein 6 (SETD6) is a mono-methyltransferase that promotes the methylation of multiple transcription factors and other proteins involved in the regulation of gene expression programs. Many of these SETD6 substrates, such as the canonical SETD6 substrate RELA, are linked to cellular pathways that are highly relevant to human health and disease. Furthermore, SETD6 regulates numerous cancerous phenotypes and guards cancer cells from apoptosis. In the past 15 years, our knowledge of SETD6 substrate methylation and the biological roles of this enzyme has grown immensely. Here we provide a comprehensive overview of SETD6 that will enhance our understanding of this enzyme's role in chromatin and in selective transcriptional control, the contextual biological roles of this enzyme, and the molecular mechanisms and pathways in which SETD6 is involved, and we highlight the major trends in the SETD6 field.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
-
- Ambler, R. P. & Rees, M. W. ε-N-methyl-lysine in bacterial flagellar protein. Nature184, 56–57 (1959). - PubMed
-
- Allfrey, V. G. & Mirsky, A. E. Structural modifications of histones and their possible role in the regulation of RNA synthesis. Science144, 559 (1964). - PubMed
-
- Rea, S. et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature406, 593–599 (2000). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

