Strigolactone insensitivity affects the hormonal homeostasis in barley
- PMID: 40102576
- PMCID: PMC11920428
- DOI: 10.1038/s41598-025-94430-2
Strigolactone insensitivity affects the hormonal homeostasis in barley
Abstract
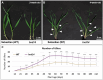
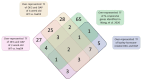
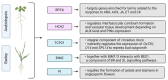
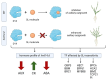
In response to environmental changes, plants continuously make architectural changes in order to optimize their growth and development. The regulation of plant branching, influenced by environmental conditions and affecting hormone balance and gene expression, is crucial for agronomic purposes due to its direct correlation with yield. Strigolactones (SL), the youngest class of phytohormones, function to shape the architecture of plants by inhibiting axillary outgrowth. Barley plants harboring the mutation in the HvDWARF14 (HvD14) gene, which encodes the SL-specific receptor, produce almost twice as many tillers as wild-type (WT) Sebastian plants. Here, through hormone profiling and comparison of transcriptomic and proteomic changes between 2- and 4-week-old plants of WT and hvd14 genotypes, we elucidate a regulatory mechanism that might affect the tillering of SL-insensitive plants. The analysis showed statistically significant increased cytokinin content and decreased auxin and abscisic acid content in 'bushy' hvd14 compared to WT, which aligns with the commonly known actions of these hormones regarding branching regulation. Further, transcriptomic and proteomic analysis revealed a set of differentially expressed genes (DEG) and abundant proteins (DAP), among which 11.6% and 14.6% were associated with phytohormone-related processes, respectively. Bioinformatics analyses then identified a series of potential SL-dependent transcription factors (TF), which may control the differences observed in the hvd14 transcriptome and proteome. Comparison to available Arabidopsis thaliana data implicates a sub-selection of these TF as being involved in the transduction of SL signal in both monocotyledonous and dicotyledonous plants.
Keywords: Hordeum vulgare; Branching; Phytohormone cross-talk; Strigolactones.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests. Ethics approval and consent to participate: All procedures were conducted in accordance to the guidelines and legislation.
Figures


References
-
- Wang, M. Molecular regulatory network of BRANCHED1 (BRC1) expression in axillary bud of Rpsa sp. in response to sugar and auxin. (Agrocampus Ouest, 2019).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

