A chromatin-focused CRISPR screen identifies USP22 as a barrier to somatic cell reprogramming
- PMID: 40102626
- PMCID: PMC11920211
- DOI: 10.1038/s42003-025-07899-y
A chromatin-focused CRISPR screen identifies USP22 as a barrier to somatic cell reprogramming
Abstract
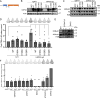
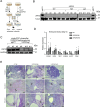
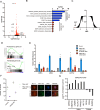
Cell-autonomous barriers to reprogramming somatic cells into induced pluripotent stem cells (iPSCs) remain poorly understood. Using a focused CRISPR-Cas9 screen, we identified Ubiquitin-specific peptidase 22 (USP22) as a key chromatin-based barrier to human iPSC derivation. Suppression of USP22 significantly enhances reprogramming efficiency. Surprisingly, this effect is likely to be independent of USP22's deubiquitinase activity or its association with the SAGA complex, as shown through module-specific knockouts, and genetic rescue experiments. USP22 is not required for iPSC derivation or maintenance. Mechanistically, USP22 loss during reprogramming downregulates fibroblast-specific genes while activating pluripotency-associated genes, including DNMT3L, LIN28A, SOX2, and GDF3. Additionally, USP22 loss enhances reprogramming efficiency under naïve stem cell conditions. These findings reveal an unrecognized role for USP22 in maintaining somatic cell identity and repressing pluripotency genes, highlighting its potential as a target to improve reprogramming efficiency.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures

References
-
- Takahashi, K. & Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell126, 663–676 (2006). - PubMed
-
- Takahashi, K. et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell131, 861–872 (2007). - PubMed
-
- David, L. & Polo, J. M. Phases of reprogramming. Stem Cell Res.12, 754–761 (2014). - PubMed
-
- Liu, X. et al. Reprogramming roadmap reveals route to human induced trophoblast stem cells. Nature586, 101–107 (2020). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

